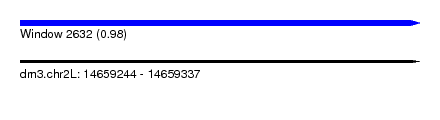
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,659,244 – 14,659,337 |
| Length | 93 |
| Max. P | 0.979832 |

| Location | 14,659,244 – 14,659,337 |
|---|---|
| Length | 93 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.75 |
| Shannon entropy | 0.40862 |
| G+C content | 0.37487 |
| Mean single sequence MFE | -29.11 |
| Consensus MFE | -15.93 |
| Energy contribution | -15.52 |
| Covariance contribution | -0.41 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.55 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.03 |
| SVM RNA-class probability | 0.979832 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14659244 93 + 23011544 -----UUUUUCUAUUAAAUGUUUUUCUCUUUCUGCGGC------GGU---------UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- -----.............(((((((((((((..((...------.))---------..))))))))))))).(((((((((..((((((((.....)))))))))))))))))--- ( -31.60, z-score = -3.53, R) >droSim1.chr2L 14407860 94 + 22036055 ----UUUUUUCUAUUAAAUGUUUUUCUCUUUCUGCAGC------GGU---------UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- ----..............(((((((((((((..((...------.))---------..))))))))))))).(((((((((..((((((((.....)))))))))))))))))--- ( -31.40, z-score = -3.42, R) >droSec1.super_3 932840 94 + 7220098 ----UUUUUUCUAUUAAAUGUUUUUCUCUUUCUGCAGC------GGU---------UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- ----..............(((((((((((((..((...------.))---------..))))))))))))).(((((((((..((((((((.....)))))))))))))))))--- ( -31.40, z-score = -3.42, R) >droYak2.chr2L 14765327 93 + 22324452 -----UUUUUCUAUUAAAUGUUUUUCUCUUUCUGCAGC------GGU---------UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- -----.............(((((((((((((..((...------.))---------..))))))))))))).(((((((((..((((((((.....)))))))))))))))))--- ( -31.40, z-score = -3.47, R) >droEre2.scaffold_4929 6406500 93 + 26641161 -----UUUUUCUAUUAAAUGUUUUUCUCUUUCUGCGGC------GGU---------UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- -----.............(((((((((((((..((...------.))---------..))))))))))))).(((((((((..((((((((.....)))))))))))))))))--- ( -31.60, z-score = -3.53, R) >droAna3.scaffold_12916 15005336 92 - 16180835 -----UUUUUCUAUUAAAUGUUUUUCUCUUUCGGCUG-----AAAAC--------GCUAGAGAGAGAGGCAGCA---ACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- -----..............((((((((((((.((((.-----...).--------)))))))))))))))((((---((((..((((((((.....)))))))))))))))).--- ( -30.20, z-score = -3.03, R) >dp4.chr4_group1 2107338 112 - 5278887 ----UUUUUUCUAUUAAUUGUUUUUCUCUUUCGACUGUUCGAAAAGCAGGUGACGGGUGAGAGGAGAAACGGUAAGCACUUGGCUACUUUUCAACUAGAAGUAGGAGUUCUUGUUG ----............((((((((((.((((((.(((((............))))).))))))))))))))))(((.((((..((((((((.....)))))))))))).))).... ( -28.10, z-score = -0.49, R) >droPer1.super_5 2052748 112 + 6813705 ----UUUUUUCUAUUAAAUGUUUUUCUCUUUCGACUGUUCGAAAAGCAGGUGACGGGUGAGAGGAGAAACGGUAAGCACUUGGCUACUUUUCAACUAGAAGUAGGAGUUCUUGUUG ----((((((((.(((..((((..((((((((((....))).)))).))).))))..))).))))))))(..((((.((((..((((((((.....)))))))))))).))))..) ( -27.40, z-score = -0.28, R) >droGri2.scaffold_15126 983073 80 - 8399593 GUUGGUUAUUCUAUUAAAUGAUUUUCU---UAGACUG------------------------AGUAAAAACA------ACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG--- .(((.(((.((((..(((....)))..---)))).))------------------------).))).((((------((((..((((((((.....)))))))))))))))).--- ( -18.90, z-score = -2.03, R) >consensus _____UUUUUCUAUUAAAUGUUUUUCUCUUUCUGCUGC______GGU_________UUAAGGAGAGAAGCAGCAGCAACUUGGCUACUUUUCAACUAGAAGUAGGAGUUGUUG___ ..................((((((((((((............................))).))))))))).((((.((((..((((((((.....)))))))))))).))))... (-15.93 = -15.52 + -0.41)

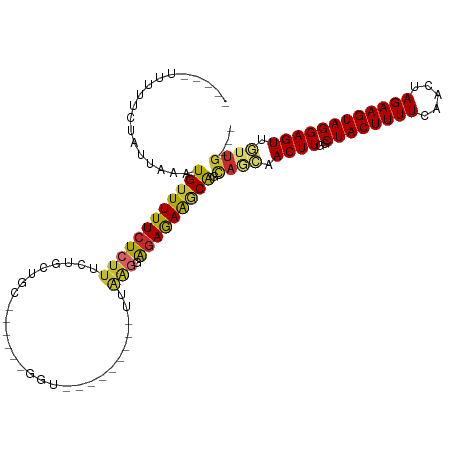
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:12 2011