| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,342,960 – 1,343,046 |
| Length | 86 |
| Max. P | 0.802355 |

| Location | 1,342,960 – 1,343,046 |
|---|---|
| Length | 86 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 64.36 |
| Shannon entropy | 0.68540 |
| G+C content | 0.49137 |
| Mean single sequence MFE | -23.01 |
| Consensus MFE | -10.42 |
| Energy contribution | -11.43 |
| Covariance contribution | 1.01 |
| Combinations/Pair | 1.45 |
| Mean z-score | -0.78 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.27 |
| SVM RNA-class probability | 0.620894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
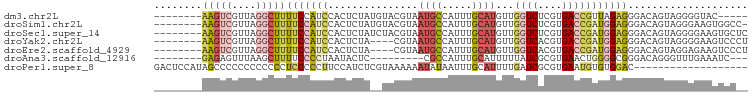
>dm3.chr2L 1342960 86 + 23011544 --------AAGUCGUUAGGCUUUUCCAUCCACUCUAUGUACGUAAUGCCAUUUGCAUGUUGGUCUCGUGACCGUUAGAGGGACAGUAGGGGUAC----- --------(((((....)))))((((.(((.(((((........((((.....))))...((((....))))..)))))))).....))))...----- ( -21.20, z-score = 0.01, R) >droSim1.chr2L 1316161 90 + 22036055 --------AAGUCGUUAGGCUUUUCCAUCCACUCUAUGUACGUAAUGCCAUUUGCAUGUUGGUCUCGUGACCGAUGGAGGGACAGUAGGGAAGUGGCC- --------.........(((((((((.(((.((((.........((((.....))))(((((((....))))))))))))))......))))).))))- ( -27.30, z-score = -0.83, R) >droSec1.super_14 1290972 91 + 2068291 --------AAGUCGUUAGGCUUUUCCAUCCACUCUAUCUACGUAAUGCCAUUUGCAUGUUGGUCUCGUGACCGAUGGAGGGACAGUAGGGGAAGUGCUC --------..........((((..((.(((.(((((((......((((.....))))...((((....)))))))))))))).....))..)))).... ( -29.70, z-score = -1.68, R) >droYak2.chr2L 1317385 87 + 22324452 --------AAGUCGUUAGGCUUUUCCAUCCACUCUA----CGUAAUGCCAUUUGCAUGUUGGUCACGUGACCGAUGGAGGGACAGUAGGGGAAGUCCCU --------.........(((((..((.(((.((((.----....((((.....))))(((((((....)))))))))))))).....))..)))))... ( -28.60, z-score = -1.39, R) >droEre2.scaffold_4929 1380514 87 + 26641161 --------AAGUCGUUAGGCUUUUCCAUCCACUCUA----CGUAAUGCCAUUUGCAUGUUGGUCACGUGACCGAUGGAGGGACAGUAGGAGAAGUCCCU --------.........(((((((((.(((.((((.----....((((.....))))(((((((....)))))))))))))).....)))))))))... ( -29.40, z-score = -2.16, R) >droAna3.scaffold_12916 4804125 79 + 16180835 --------GAGAGUUUAAGCUUUUCCCCUAAUACUC---------CGCCAUUUGCAUUUUUAUCGCGUGAACUGGGGCGGGACAGGGUUUGAAAUC--- --------..((.((((((((((((((((.....((---------(((.((..........)).))).)).....)).)))).)))))))))).))--- ( -18.20, z-score = 0.23, R) >droPer1.super_8 1047157 80 + 3966273 GACUCCAUAGCCCCCCCCCCCCUCCCCCUUCCAUCUCGUAAAAAAUAUAAUUUGCAUUUUGAUCGCGUGAAUGUGUGGAC------------------- ...((((((............................(((((........))))).......((....))...)))))).------------------- ( -6.70, z-score = 0.37, R) >consensus ________AAGUCGUUAGGCUUUUCCAUCCACUCUA__UACGUAAUGCCAUUUGCAUGUUGGUCUCGUGACCGAUGGAGGGACAGUAGGGGAAGUCC__ ........(((((....)))))(((((((...............((((.....))))...((((....))))))))))).................... (-10.42 = -11.43 + 1.01)


| Location | 1,342,960 – 1,343,046 |
|---|---|
| Length | 86 |
| Sequences | 7 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 64.36 |
| Shannon entropy | 0.68540 |
| G+C content | 0.49137 |
| Mean single sequence MFE | -20.56 |
| Consensus MFE | -10.13 |
| Energy contribution | -10.73 |
| Covariance contribution | 0.60 |
| Combinations/Pair | 1.43 |
| Mean z-score | -0.81 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.74 |
| SVM RNA-class probability | 0.802355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1342960 86 - 23011544 -----GUACCCCUACUGUCCCUCUAACGGUCACGAGACCAACAUGCAAAUGGCAUUACGUACAUAGAGUGGAUGGAAAAGCCUAACGACUU-------- -----.........(((((((((((..((((....))))...((((.....))))........))))).))))))..(((.(....).)))-------- ( -22.20, z-score = -1.97, R) >droSim1.chr2L 1316161 90 - 22036055 -GGCCACUUCCCUACUGUCCCUCCAUCGGUCACGAGACCAACAUGCAAAUGGCAUUACGUACAUAGAGUGGAUGGAAAAGCCUAACGACUU-------- -(((...((((.....(((((((....((((....))))...((((.....))))..........))).))))))))..))).........-------- ( -22.60, z-score = -1.20, R) >droSec1.super_14 1290972 91 - 2068291 GAGCACUUCCCCUACUGUCCCUCCAUCGGUCACGAGACCAACAUGCAAAUGGCAUUACGUAGAUAGAGUGGAUGGAAAAGCCUAACGACUU-------- ..((..((((......(((((((.(((((((....))))...((((.....))))......))).))).))))))))..))..........-------- ( -23.10, z-score = -1.50, R) >droYak2.chr2L 1317385 87 - 22324452 AGGGACUUCCCCUACUGUCCCUCCAUCGGUCACGUGACCAACAUGCAAAUGGCAUUACG----UAGAGUGGAUGGAAAAGCCUAACGACUU-------- .(((((..........)))))((((((((((....))))...((((.....))))....----.......)))))).(((.(....).)))-------- ( -25.50, z-score = -1.41, R) >droEre2.scaffold_4929 1380514 87 - 26641161 AGGGACUUCUCCUACUGUCCCUCCAUCGGUCACGUGACCAACAUGCAAAUGGCAUUACG----UAGAGUGGAUGGAAAAGCCUAACGACUU-------- .(((((..........)))))((((((((((....))))...((((.....))))....----.......)))))).(((.(....).)))-------- ( -25.50, z-score = -1.52, R) >droAna3.scaffold_12916 4804125 79 - 16180835 ---GAUUUCAAACCCUGUCCCGCCCCAGUUCACGCGAUAAAAAUGCAAAUGGCG---------GAGUAUUAGGGGAAAAGCUUAAACUCUC-------- ---.........(((((.((((((...(....)(((.......)))....))))---------).)...))))).................-------- ( -16.50, z-score = -0.23, R) >droPer1.super_8 1047157 80 - 3966273 -------------------GUCCACACAUUCACGCGAUCAAAAUGCAAAUUAUAUUUUUUACGAGAUGGAAGGGGGAGGGGGGGGGGGGCUAUGGAGUC -------------------((((.(...(((.(.(..((....(.((..(..((.....))..)..)).)...))..).))))..)))))......... ( -8.50, z-score = 2.18, R) >consensus __GGACUUCCCCUACUGUCCCUCCAUCGGUCACGAGACCAACAUGCAAAUGGCAUUACGUA__UAGAGUGGAUGGAAAAGCCUAACGACUU________ .....................((((((((((....))))...((((.....))))...............))))))....................... (-10.13 = -10.73 + 0.60)

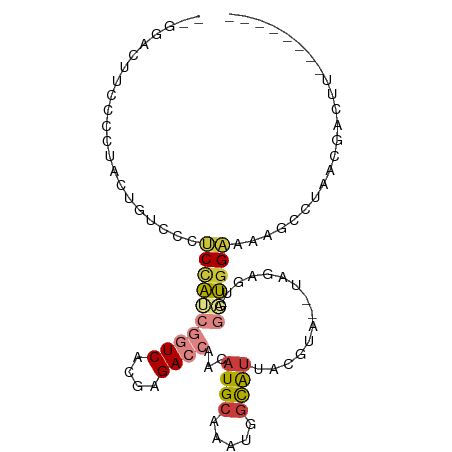

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:22 2011