
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,639,002 – 14,639,118 |
| Length | 116 |
| Max. P | 0.814690 |

| Location | 14,639,002 – 14,639,118 |
|---|---|
| Length | 116 |
| Sequences | 8 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.65 |
| Shannon entropy | 0.42322 |
| G+C content | 0.53504 |
| Mean single sequence MFE | -26.38 |
| Consensus MFE | -16.40 |
| Energy contribution | -16.65 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.814690 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
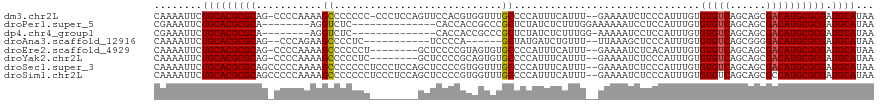
>dm3.chr2L 14639002 116 - 23011544 CAAAAUUCUGCACGCGCAG-CCCCAAAAGCCCCCCC-CCCUCCAGUUCCACGUGGUUUGCCCCAUUUCAUUU--GAAAAUCUCCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........((((((((((.-................-...........((((.((....)).).((((....--))))...........)))((((......)))))))))).))))... ( -23.70, z-score = -1.01, R) >droPer1.super_5 2037034 98 - 6813705 CGAAAUUCUGCACGCGCA--------AGGUCUC--------------CACCACCGCCCGCUCUAUCUCUUUGGAAAAAAUCCUCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........((((((((((--------.(((...--------------.)))..(((..(((....(.(..((((........))))...).)....))).)))...)))))).))))... ( -27.90, z-score = -1.96, R) >dp4.chr4_group1 2091515 97 + 5278887 CGAAAUUCUGCACGCGCA--------AGGUCUC--------------CACCACCGCCCGCUCUAUCUCUUUGG-AAAAAUCCUCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........((((((((((--------.(((...--------------.)))..(((..(((....(.(..(((-(.......))))...).)....))).)))...)))))).))))... ( -27.60, z-score = -1.80, R) >droAna3.scaffold_12916 14994510 99 + 16180835 CAAAAUUCUGCACGCGCAG--CCCAGAAGCCCCUC-----------UCCCCA------GCUAUGAUCUGUUU--UUAAAGCUCCCAUUUGUGUGUCAGCGGGGACAUGCGCGAUGCAUAA ........(((((((((((--(......)).....-----------((((((------((........))).--.....(((..(((....)))..))))))))..)))))).))))... ( -30.30, z-score = -1.71, R) >droEre2.scaffold_4929 6395097 109 - 26641161 CAAAAUUCUGCACGCGCAG-CCCCAAAAGCCCCCCU--------GCUCCCCGUAGUGUGCCCCAUUUCAUUU--GAAAAUCUCACAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........(((((((((((-(.......))...(((--------(((..((..((((((.....((((....--))))....))))))...).)..))))).)...)))))).))))... ( -27.50, z-score = -2.22, R) >droYak2.chr2L 14753967 109 - 22324452 CAAAAUUCUGCACGCGCAG-CCCCAAAAGCCCCCUC--------GCUCCCCGCAGUGUGCCCCAUUUCAUUU--GAAAAUCUCCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........(((((((((((-(.......))....((--------((((..((((..(((.....((((....--))))......)))...))))...).)))))..)))))).))))... ( -25.20, z-score = -1.41, R) >droSec1.super_3 921834 118 - 7220098 CAAAAUUCUGCACGCGCAGCCCCCAAAAGCCCCCCCUCCCUCCAGCUCCCCGUGGUUUGCCCCAUUUCAUUU--GAAAAUCUCCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........(((((((((((.(((....(((..............)))....).)).))))....((((....--))))...........(((((((......)))))))))).))))... ( -23.64, z-score = -0.94, R) >droSim1.chr2L 14396739 118 - 22036055 CAAAAUUCUGCACGCGCAGCCCCCAAAAGCCCCCCCUCCCUCCAGCUCCCCGUGGUUUGCCCCAUUUCAUUU--GAAAAUCUCCCAUUUGUGUGUCAGCAGCGCCAUGCGCGAUGCAUAA ........(((((((((.((........)).....................(((((((((..(((..((..(--(.........))..)).)))...)))).)))))))))).))))... ( -25.20, z-score = -1.52, R) >consensus CAAAAUUCUGCACGCGCAG_CCCCAAAAGCCCCCCC________GCUCCCCGUGGUUUGCCCCAUUUCAUUU__GAAAAUCUCCCAUUUGUGUGUCAGCAGCGACAUGCGCGAUGCAUAA ........(((((((((...........................................................((((.....))))..(((((......)))))))))).))))... (-16.40 = -16.65 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:09 2011