
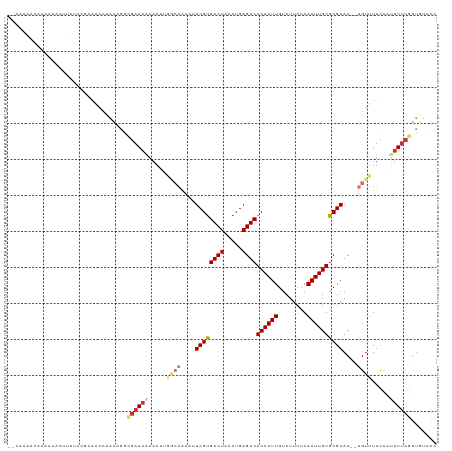
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,638,439 – 14,638,554 |
| Length | 115 |
| Max. P | 0.884044 |

| Location | 14,638,439 – 14,638,554 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 74.95 |
| Shannon entropy | 0.50872 |
| G+C content | 0.34423 |
| Mean single sequence MFE | -20.66 |
| Consensus MFE | -13.15 |
| Energy contribution | -14.47 |
| Covariance contribution | 1.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.884044 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14638439 115 + 23011544 --AAAAAUAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC--UGUUUUCCAUUUUCGGUGUCCC --..................((((((..((((((.((.....)).))....(((((((((.....))))((((((........))))))))))))--)))......))))))....... ( -20.80, z-score = -1.13, R) >droSim1.chr2L 14396175 115 + 22036055 --AAAUAAAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC--UGUUUUCCAUUUUCGGUGUCCC --..................((((((..((((((.((.....)).))....(((((((((.....))))((((((........))))))))))))--)))......))))))....... ( -20.80, z-score = -1.13, R) >droSec1.super_3 921271 117 + 7220098 AAAUAAAAAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGCGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC--UGUUUUCCAUUUUCGGUGUCCC ....................((((((..((((((.((.....)).))....(((((((((.....))))((((((........))))))))))))--)))......))))))....... ( -23.30, z-score = -1.85, R) >droYak2.chr2L 14753392 105 + 22324452 ------------AAAAUUUACGAAAACUACAGGCGAAAAAAAUGGACAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC--UGUUUUCCAUUUUCGGUGUCCC ------------...................((((...((((((((.((..(((((((((.....))))((((((........))))))))))).--..)).))))))))...)))).. ( -23.20, z-score = -1.88, R) >droEre2.scaffold_4929 6394538 105 + 26641161 ------------AAAUUUUACAAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC--UGUUUUCCAUUUUCGGUGUGCC ------------...................((((....(((.(((.(((.(((((((((.....))))((((((........))))))))))).--..))).))).))).....)))) ( -19.70, z-score = -0.69, R) >dp4.chr4_group1 2091032 94 - 5278887 --AACACCGAAAAAUUGUU---------------GAAAAAAA--------AAUCGUGACUAAAUGAGUCACAAUUGUCUUCCAAAUUGUGCGAACACUGUGUUUUUCUUUCAGUUCCCC --..........(((((..---------------((((((..--------....((((((.....))))))....((.(((((.....)).))).))....))))))...))))).... ( -15.20, z-score = -0.77, R) >droPer1.super_5 2036530 109 + 6813705 --AACACCGAAAAAUUGUUUAAAAAAAAGUCGGUGAAAAAAA--------AAUCGUGACUAAAUGAGUCACAAUUGUCUUCCAAAUUGUGCGAACACUGUGUUUUUCUUUCAGUUCCCC --..((((((....((.......))....)))))).......--------..((((((((.....))))((((((........))))))))))..((((...........))))..... ( -19.60, z-score = -1.28, R) >droWil1.scaffold_180764 3012752 115 + 3949147 --GAAACCGAAACACAGAAACAGAAACAGUAACGGAACGAAAACAACAAAAAUCGUGACUUAAUGAGUCACAAUUGUCUUCCAAAUUGUGCGAAC--UGUUUUCGUUUUAAGACACACC --..........................((...((((((((((((.......((((......))))(.(((((((........))))))))....--))))))))))))...))..... ( -22.70, z-score = -1.07, R) >consensus __AAAAACAAAAAACUUUUACGAAAACUACAGGCGAAAAAAAUUGGCAAAAUUCGUGACUAAAUGAGUCACAAUUGUCUUCUAAAUUGUGCGAAC__UGUUUUCCAUUUUCGGUGUCCC .................................((((((.....((((....((((((((.....))))((((((........))))))))))....)))).....))))))....... (-13.15 = -14.47 + 1.33)


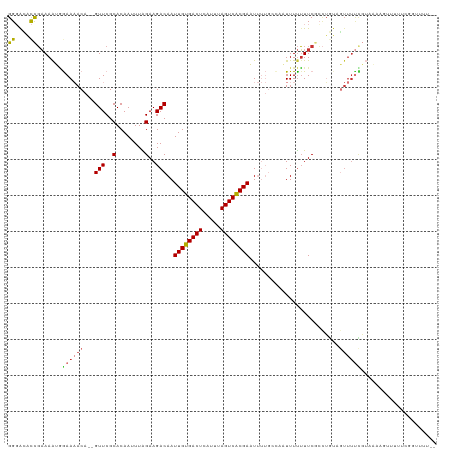
| Location | 14,638,439 – 14,638,554 |
|---|---|
| Length | 115 |
| Sequences | 8 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 74.95 |
| Shannon entropy | 0.50872 |
| G+C content | 0.34423 |
| Mean single sequence MFE | -23.03 |
| Consensus MFE | -12.46 |
| Energy contribution | -12.90 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.12 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.15 |
| SVM RNA-class probability | 0.567738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14638439 115 - 23011544 GGGACACCGAAAAUGGAAAACA--GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCACGAAUUUUGCCAAUUUUUUUCGCCUGUAGUUUUCGUAAAAGUUUUUUAUUUUU-- ((....))(((((((((((((.--((....))..((((((((((..((((((((.....)))))))).....((.((.....)).)).....)))))).)))).)))))))))))))-- ( -25.20, z-score = -1.81, R) >droSim1.chr2L 14396175 115 - 22036055 GGGACACCGAAAAUGGAAAACA--GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCACGAAUUUUGCCAAUUUUUUUCGCCUGUAGUUUUCGUAAAAGUUUUUUUUAUUU-- ((....))......(((((((.--((....))..((((((((((..((((((((.....)))))))).....((.((.....)).)).....)))))).)))).)))))))......-- ( -19.10, z-score = 0.20, R) >droSec1.super_3 921271 117 - 7220098 GGGACACCGAAAAUGGAAAACA--GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCGCGAAUUUUGCCAAUUUUUUUCGCCUGUAGUUUUCGUAAAAGUUUUUUUUUUAUUU ((....)).......(((((((--((((((((((((........))))))((((.....)))))))))))..((.((.....)).)).....))))))(((((((.....))))))).. ( -22.40, z-score = -0.52, R) >droYak2.chr2L 14753392 105 - 22324452 GGGACACCGAAAAUGGAAAACA--GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCACGAAUUUUGUCCAUUUUUUUCGCCUGUAGUUUUCGUAAAUUUU------------ (((.....(((((((((....(--(((((.((((((........))))))((((.....)))).))))))...)))))))))....)))..................------------ ( -22.00, z-score = -1.11, R) >droEre2.scaffold_4929 6394538 105 - 26641161 GGCACACCGAAAAUGGAAAACA--GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCACGAAUUUUGCCAAUUUUUUUCGCCUGUAGUUUUUGUAAAAUUU------------ (((.((.......))(((((.(--(((.(((.(((((.(....)....((((((.....))))))))))).))).)))).))))))))...................------------ ( -18.70, z-score = -0.27, R) >dp4.chr4_group1 2091032 94 + 5278887 GGGGAACUGAAAGAAAAACACAGUGUUCGCACAAUUUGGAAGACAAUUGUGACUCAUUUAGUCACGAUU--------UUUUUUUC---------------AACAAUUUUUCGGUGUU-- .....(((((((((........(((....)))...((((((((.((((((((((.....))))))))))--------..))))))---------------))...)))))))))...-- ( -26.30, z-score = -3.03, R) >droPer1.super_5 2036530 109 - 6813705 GGGGAACUGAAAGAAAAACACAGUGUUCGCACAAUUUGGAAGACAAUUGUGACUCAUUUAGUCACGAUU--------UUUUUUUCACCGACUUUUUUUUAAACAAUUUUUCGGUGUU-- .....(((((((((........(((....)))....(((((((.((((((((((.....))))))))))--------..)))))))...................)))))))))...-- ( -25.40, z-score = -1.76, R) >droWil1.scaffold_180764 3012752 115 - 3949147 GGUGUGUCUUAAAACGAAAACA--GUUCGCACAAUUUGGAAGACAAUUGUGACUCAUUAAGUCACGAUUUUUGUUGUUUUCGUUCCGUUACUGUUUCUGUUUCUGUGUUUCGGUUUC-- ............((((((((((--(((..(........)..)))((((((((((.....)))))))))).....))))))))))(((..((...............))..)))....-- ( -25.16, z-score = -0.68, R) >consensus GGGACACCGAAAAUGGAAAACA__GUUCGCACAAUUUAGAAGACAAUUGUGACUCAUUUAGUCACGAAUUUUGCCAAUUUUUUUCGCCUGUAGUUUUCGUAAAAGUUUUUCGGUUUU__ ((....)).......((((((...(((..(........)..)))..((((((((.....)))))))).........................))))))..................... (-12.46 = -12.90 + 0.44)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:08 2011