
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,602,790 – 14,602,904 |
| Length | 114 |
| Max. P | 0.723898 |

| Location | 14,602,790 – 14,602,904 |
|---|---|
| Length | 114 |
| Sequences | 11 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 73.68 |
| Shannon entropy | 0.58953 |
| G+C content | 0.59361 |
| Mean single sequence MFE | -41.60 |
| Consensus MFE | -16.04 |
| Energy contribution | -17.52 |
| Covariance contribution | 1.48 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.62 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723898 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
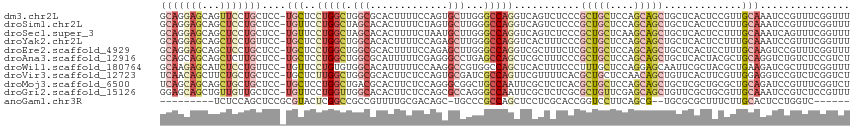
>dm3.chr2L 14602790 114 + 23011544 GCAGGAGCAGUUCCUGCUCC-UGCUCCUGGCUGGCGCACUUUUCCAGUGCUUGGGCCAGGUCAGUCUCCCGCUGCUCCAGCAGCUGCUCACUCCGUUGCAAAUCCGUUUCGGUUU ((((((((((...)))))))-))).(((((((.(.(((((.....))))).).)))))))..........(((((....)))))........(((..((......))..)))... ( -52.40, z-score = -4.31, R) >droSim1.chr2L 14354674 114 + 22036055 GCAGGAGCAGCUCCUGCUCC-UGUUCCUGGCUAGCACACUUUUCUAGUGCUUGGGCCAGGUCAGUCUCCCGCUGCUCCAGCAGCUGCUCACUCCUUUGCAAAUCCGUUUCGGUUU ((((((((((...)))))))-))).((((((((((((.........)))))..)))))))........(((((((....)))))(((..........)))..........))... ( -45.70, z-score = -3.44, R) >droSec1.super_3 876494 114 + 7220098 GCAGGAGCAGCUCCUGCUCC-UGUUCCUGGCUAGCACACUUUUCUAAUGCUUGGGCCAGGUCAGUCUCCCGCUGCUCAAGCAGCUGCUCACUCCUUUGCAAAUCAGUUUCGGUUU ((((((((((...)))))))-))).(((((((((((...........))))..)))))))........(((((((....)))))(((..........)))..........))... ( -41.90, z-score = -2.40, R) >droYak2.chr2L 14716796 114 + 22324452 GCAGGAGCAGCUCCUGUUCC-UGCUCCUGGCUGGCACACUUUUCCAGAGCUUGGGCCAGGUCACUUUCCCGCUGCUCCAGCAGCUGCUCACUCCUUUGCAAAUCCGUUUCGGUUU ((((((((((...)))))))-))).((((((((((.............)))..)))))))........(((((((....)))))(((..........)))..........))... ( -41.12, z-score = -1.51, R) >droEre2.scaffold_4929 6359483 114 + 26641161 GCAGGAGCAGCUCCUGCUCC-UGCUCCUGGCUGGCGCACUUUUCCAGAGCUUGGGCCAGGUCGCUUUCUCGCUGCUCCAGCAGCUGCUCACUCCUUUGCAAGUCCGUUUCGGUUU ((((((((((...)))))))-))).(((((((.(.((.((.....)).)).).)))))))..........(((((....)))))(((..........)))...(((...)))... ( -45.10, z-score = -1.16, R) >droAna3.scaffold_12916 14961262 114 - 16180835 GCAGCAGCAGCUCUUGCUCC-UGCUCCUGGCUGGCGCAUUUUUCGAGGGCCUGAGCCAGCUCGCUUUCCCGCUGCUCCAGCAGCUGCUCACUACGCUGCAGGUCUGUCUCCGUCU ((((((((((((...((...-.)).....(((((.(((......((((((..(((....)))))))))....))).))))))))))))......))))).((.......)).... ( -41.50, z-score = 0.69, R) >droWil1.scaffold_180764 2963720 114 + 3949147 GCAAGAGCAUCUCCUGUUCC-UGUUCCUGUGUGGCACAUUUUUCCAAGGCCGUGGCCAGCUCACUUUCCCUUUGCUCCAGGAGCAAUUCGCUACGCUGAAGAUCGCUUUCGGUUU (((.(((((.....))))).-)))((..(((((((............(((....)))..............((((((...))))))...))))))).))((((((....)))))) ( -30.30, z-score = -0.00, R) >droVir3.scaffold_12723 2492218 114 + 5802038 UCAACAGCUUCUGCUGCUCC-UGCUCUUGGCUGGCGCACUUCUCCAGUGCGAUCGCCAGUUCGUUUUCACGCUGCUCCAACAGCUGUUCACUUCGUUGGAGGUCCGUCUCGGUCU ....((((....))))((((-.((..(((((...((((((.....))))))...)))))...((...((.((((......))))))...))...)).))))(.(((...))).). ( -35.20, z-score = -1.07, R) >droMoj3.scaffold_6500 15677791 114 + 32352404 UCAGCAGCAGCUGCUGCUCC-UGCUCCUGGCUGACGCACUUCUCCAGGGCGGCUGCCAAUUCGCUCUCACGCUGCUCCAGCAGCUGCUCGCUGCGCUGCAGAUCCGUUUCGGUCU ..(((((.(((....))).)-))))...((((((.((...(((.(((.(((((.((......))......(((((....))))).....))))).))).)))...)).)))))). ( -45.50, z-score = -0.41, R) >droGri2.scaffold_15126 937174 114 - 8399593 GGAGCAGCUGUUGUUGCUCC-UGUUCCUGGUUGGCACACUUCUCCAGCGCCAGGGCCAAUUCGCUCUCGCGCUGUUCGAGCAGCUGUUCGCUGCGUUGCAAAUCCGUCUCCGUUU ((((((((....))))))))-.......((...(((.((.....((((((.(((((......))))).)))))).....(((((.....))))))))))....)).......... ( -46.30, z-score = -2.95, R) >anoGam1.chr3R 44806933 97 + 53272125 ---------UCUCCAGCUCCGCGUACUCGGCCGCCGUUUUGCGACAGC-UGCCCGCCAGCUCCUCGCACCGGUCCUUCAGCG--UGCGCGCUUUCUUGCACUCCUGGUC------ ---------...((((....((((((.(....((((...(((((.(((-((.....)))))..))))).))))......).)--)))))((......))....))))..------ ( -32.60, z-score = -1.26, R) >consensus GCAGGAGCAGCUCCUGCUCC_UGCUCCUGGCUGGCGCACUUUUCCAGUGCUUGGGCCAGGUCACUCUCCCGCUGCUCCAGCAGCUGCUCACUCCGUUGCAAAUCCGUUUCGGUUU (((((((...)))))))....(((..(((((.(((.............)))...)))))...........(((((....))))).............)))............... (-16.04 = -17.52 + 1.48)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:41:01 2011