
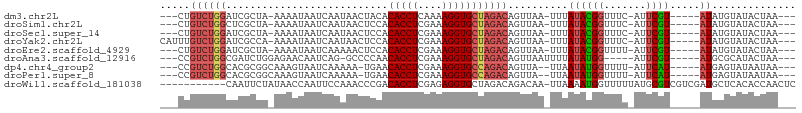
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 92,599 – 92,691 |
| Length | 92 |
| Max. P | 0.959955 |

| Location | 92,599 – 92,691 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | forward |
| Mean pairwise identity | 76.60 |
| Shannon entropy | 0.45093 |
| G+C content | 0.37345 |
| Mean single sequence MFE | -19.83 |
| Consensus MFE | -8.84 |
| Energy contribution | -9.05 |
| Covariance contribution | 0.21 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.98 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.723271 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
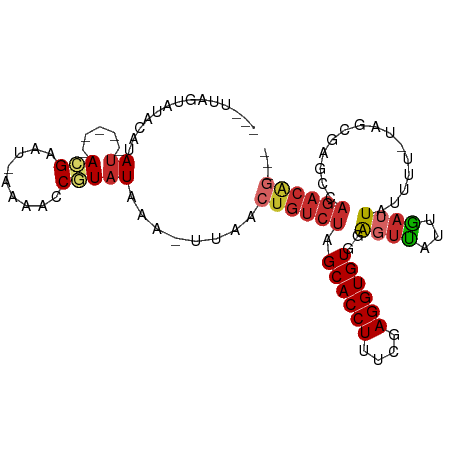
>dm3.chr2L 92599 92 + 23011544 ---CUGUCUGGAUCGCUA-AAAAUAAUCAAUAACUACACACCUCAAAAGGUGCUAGACAGUUAA-UUUAUACGGUUUC-AUUCGU-----AUAUGUAUACUAA--- ---((((((((.......-....((.....))......(((((....)))))))))))))....-..(((((((....-..))))-----)))..........--- ( -18.90, z-score = -2.56, R) >droSim1.chr2L 93029 92 + 22036055 ---CUGUCUGGCUCGCUA-AAAAUAAUCAAUAACUCCACACCUCGAAAGGUGCUAGACAGUUAA-UUUAUACGGUUUC-AUUCGU-----AUAUGUAUACUAA--- ---(((((((((......-....((.....)).......((((....)))))))))))))....-..(((((((....-..))))-----)))..........--- ( -19.70, z-score = -2.52, R) >droSec1.super_14 91713 92 + 2068291 ---CUGUCUGGAUCGCUA-AAAAUAAUCAAUAACUCCACACCUCGAAAGGUGCUAGACAGUUAA-UUUAUACGGUUUC-AUUCGU-----AUAUGUAUACUAA--- ---((((((((.......-....((.....))......(((((....)))))))))))))....-..(((((((....-..))))-----)))..........--- ( -18.90, z-score = -2.12, R) >droYak2.chr2L 81974 95 + 22324452 CAUUUGUCUGGAUCGCCA-AAAAUAAUCAAUAACUCCACACCUCGAAAGGUGCUAGACAGUUAA-UUUAUACGGUUUC-AUUCGU-----AUAUGUAUACUAA--- .....((((((....)).-...................(((((....)))))..))))(((...-..(((((((....-..))))-----))).....)))..--- ( -17.90, z-score = -1.57, R) >droEre2.scaffold_4929 120604 92 + 26641161 ---CUGUCUGGAUCGCUA-AAAAUAAUCAAAAACUCCACACCUCGAAAGGUGCUAGACAGUUAA-UUUAUACGGUUUU-AUUCGU-----AUAUGUAUACUAA--- ---((((((((.......-...................(((((....)))))))))))))....-..(((((((....-..))))-----)))..........--- ( -18.76, z-score = -2.18, R) >droAna3.scaffold_12916 2082213 89 + 16180835 ---CCGUCUGGCGAUCUGGAGAACAAUCAG-GCCCCAACACCUCGAAAGGUGCUAGACAGUUAAUUUUAUAUGG-----AUUCGU-----AUGCGCAUACUAA--- ---..((((((((..((((.......))))-..).....((((....)))))))))))(((......((((((.-----...)))-----))).....)))..--- ( -20.10, z-score = -0.22, R) >dp4.chr4_group2 1197234 91 - 1235136 ---CCGUCUGGCACGCGGCAAAGUAAUCAAAAA-UGAACACCUCGAAAGGUGCCAGACAGUUA--UUAAUAUGGUUUU-AUUCAU-----AUGAGUAUAAUAA--- ---..(((((((((((......)).........-.........(....)))))))))).((((--(..((((((....-..))))-----))....)))))..--- ( -25.10, z-score = -3.38, R) >droPer1.super_8 3857700 91 + 3966273 ---CCGUCUGGCACGCGGCAAAGUAAUCAAAAA-UGAACACCUCGAAAGGUGCCAGACAGUUA--UUAAUAUGGUUUU-AUUCAU-----AUGAGUAUAAUAA--- ---..(((((((((((......)).........-.........(....)))))))))).((((--(..((((((....-..))))-----))....)))))..--- ( -25.10, z-score = -3.38, R) >droWil1.scaffold_181038 566164 94 - 637489 -----------CAAUUCUAUAACCAAUUCCAAACCCGACACCUCGAGAGGUGCUAGACAGACAA-UUAAAAUGGUUUUUAUGCGUCGUCGAUGCUCACACCAACUC -----------.........................(.(((((....))))))...........-......((((......(((((...)))))....)))).... ( -14.00, z-score = 0.09, R) >consensus ___CUGUCUGGCUCGCUA_AAAAUAAUCAAAAACUCCACACCUCGAAAGGUGCUAGACAGUUAA_UUUAUACGGUUUC_AUUCGU_____AUAUGUAUACUAA___ .....((((((...........................(((((....)))))))))))............((((.......))))..................... ( -8.84 = -9.05 + 0.21)
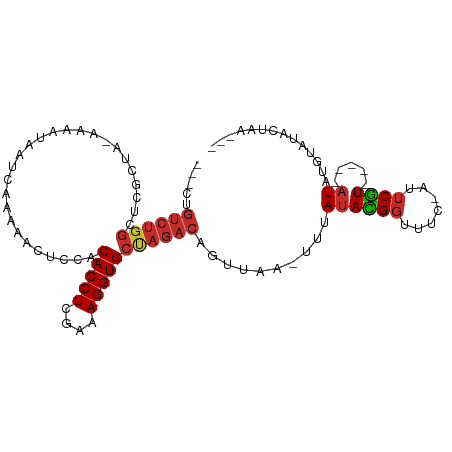
| Location | 92,599 – 92,691 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 76.60 |
| Shannon entropy | 0.45093 |
| G+C content | 0.37345 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -9.06 |
| Energy contribution | -9.62 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.37 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959955 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 92599 92 - 23011544 ---UUAGUAUACAUAU-----ACGAAU-GAAACCGUAUAAA-UUAACUGUCUAGCACCUUUUGAGGUGUGUAGUUAUUGAUUAUUUU-UAGCGAUCCAGACAG--- ---..........(((-----(((...-.....))))))..-....((((((.((((((....))))))...((((...........-)))).....))))))--- ( -20.50, z-score = -2.34, R) >droSim1.chr2L 93029 92 - 22036055 ---UUAGUAUACAUAU-----ACGAAU-GAAACCGUAUAAA-UUAACUGUCUAGCACCUUUCGAGGUGUGGAGUUAUUGAUUAUUUU-UAGCGAGCCAGACAG--- ---..........(((-----(((...-.....))))))..-....((((((.((((((....))))))((.((((...........-))))...))))))))--- ( -23.10, z-score = -2.90, R) >droSec1.super_14 91713 92 - 2068291 ---UUAGUAUACAUAU-----ACGAAU-GAAACCGUAUAAA-UUAACUGUCUAGCACCUUUCGAGGUGUGGAGUUAUUGAUUAUUUU-UAGCGAUCCAGACAG--- ---..........(((-----(((...-.....))))))..-....((((((.((((((....))))))(((.(..((((......)-)))..))))))))))--- ( -23.30, z-score = -3.04, R) >droYak2.chr2L 81974 95 - 22324452 ---UUAGUAUACAUAU-----ACGAAU-GAAACCGUAUAAA-UUAACUGUCUAGCACCUUUCGAGGUGUGGAGUUAUUGAUUAUUUU-UGGCGAUCCAGACAAAUG ---..........(((-----(((...-.....))))))..-.....(((((.((((((....))))))(((.(..(..(......)-..)..))))))))).... ( -21.10, z-score = -1.74, R) >droEre2.scaffold_4929 120604 92 - 26641161 ---UUAGUAUACAUAU-----ACGAAU-AAAACCGUAUAAA-UUAACUGUCUAGCACCUUUCGAGGUGUGGAGUUUUUGAUUAUUUU-UAGCGAUCCAGACAG--- ---..........(((-----(((...-.....))))))..-....((((((.((((((....))))))(((....((((......)-)))...)))))))))--- ( -22.70, z-score = -3.13, R) >droAna3.scaffold_12916 2082213 89 - 16180835 ---UUAGUAUGCGCAU-----ACGAAU-----CCAUAUAAAAUUAACUGUCUAGCACCUUUCGAGGUGUUGGGGC-CUGAUUGUUCUCCAGAUCGCCAGACGG--- ---...((((....))-----))....-----..............(((((((((((((....)))))))..(((-..((((........)))))))))))))--- ( -22.40, z-score = -0.81, R) >dp4.chr4_group2 1197234 91 + 1235136 ---UUAUUAUACUCAU-----AUGAAU-AAAACCAUAUUAA--UAACUGUCUGGCACCUUUCGAGGUGUUCA-UUUUUGAUUACUUUGCCGCGUGCCAGACGG--- ---((((((.....((-----(((...-.....))))))))--)))(((((((((((....(((((((.(((-....))).)))))))....)))))))))))--- ( -30.40, z-score = -5.96, R) >droPer1.super_8 3857700 91 - 3966273 ---UUAUUAUACUCAU-----AUGAAU-AAAACCAUAUUAA--UAACUGUCUGGCACCUUUCGAGGUGUUCA-UUUUUGAUUACUUUGCCGCGUGCCAGACGG--- ---((((((.....((-----(((...-.....))))))))--)))(((((((((((....(((((((.(((-....))).)))))))....)))))))))))--- ( -30.40, z-score = -5.96, R) >droWil1.scaffold_181038 566164 94 + 637489 GAGUUGGUGUGAGCAUCGACGACGCAUAAAAACCAUUUUAA-UUGUCUGUCUAGCACCUCUCGAGGUGUCGGGUUUGGAAUUGGUUAUAGAAUUG----------- ..(((((((....)))))))..........(((((((((((-...((((....((((((....)))))))))).)))))).))))).........----------- ( -24.60, z-score = -1.00, R) >consensus ___UUAGUAUACAUAU_____ACGAAU_AAAACCGUAUAAA_UUAACUGUCUAGCACCUUUCGAGGUGUGGAGUUAUUGAUUAUUUU_UAGCGAGCCAGACAG___ ..............................................((((((.((((((....))))))..((((...))))...............))))))... ( -9.06 = -9.62 + 0.56)
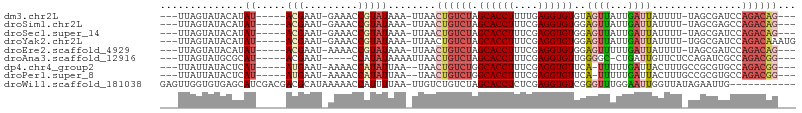
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:05:06 2011