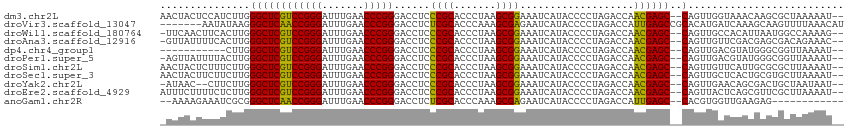
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,598,834 – 14,598,944 |
| Length | 110 |
| Max. P | 0.976371 |

| Location | 14,598,834 – 14,598,944 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 79.05 |
| Shannon entropy | 0.47958 |
| G+C content | 0.53087 |
| Mean single sequence MFE | -35.64 |
| Consensus MFE | -30.01 |
| Energy contribution | -28.97 |
| Covariance contribution | -1.04 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.57 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.730508 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
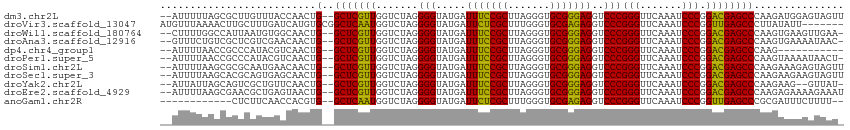
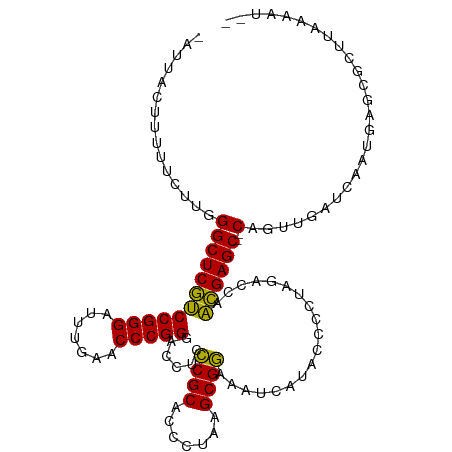
>dm3.chr2L 14598834 110 + 23011544 --AUUUUUAGCGCUUGUUUACCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAUGGAGUAGUU --.......((((((((((((((((..--....))))))...(((((.(((..(((((.......))(((....)))))).))).)))))))))))))((.....)).)).... ( -37.20, z-score = -0.49, R) >droVir3.scaffold_13047 2079034 107 - 19223366 AUGUUUAAAACUUGCUUUGAUCAUGUGCGGCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCUUAUAUU------- (((.(((((......))))).)))....((((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))........------- ( -33.60, z-score = -1.11, R) >droWil1.scaffold_180764 2954693 109 + 3949147 --CUUUUGGCCAUUAAUGUGGCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGUGAAGUUGAA- --..(((((((((....)))))....(--(((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))))))..........- ( -40.40, z-score = -1.20, R) >droAna3.scaffold_12916 14958224 109 - 16180835 --GUUUCUGUCGCUCGUCGAACAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGUGAAAAUAAC- --.((((....(((((((((.((((..--....)))).))..(((((.(((..(((((.......))(((....)))))).))).))))).)))))))......)))).....- ( -38.20, z-score = -0.98, R) >dp4.chr4_group1 2039688 99 - 5278887 --AUUUUAACCGCCCAUACGUCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAG----------- --........................(--(((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))....----------- ( -32.40, z-score = 0.24, R) >droPer1.super_5 1983253 109 + 6813705 --AUUUUAACCGCCCAUACGUCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGUAAAAUAACU- --(((((((((((((.((..(((((..--....)))))..)).)))).....((((((.......))))))))).(((((.......)))))...........))))))....- ( -33.20, z-score = 0.25, R) >droSim1.chr2L 14350385 110 + 22036055 --AUUUUAAGCGCGCAAUGAACAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAAAGAGUAGUU --.......((((.((((((.(....)--..)))))).((((((((........)).))))))))))(((..((((.(((.......)))))))...))).............. ( -33.70, z-score = 0.11, R) >droSec1.super_3 872538 110 + 7220098 --AUUUUAAGCACGCAGUGAGCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAAGAAGUAGUU --(((((..((((.((((((((.....--)))))))).((((((((........)).))))))))))(((..((((.(((.......)))))))...))).....))))).... ( -37.90, z-score = -1.03, R) >droYak2.chr2L 14713281 107 + 22324452 --AUUAUUAGCAGUCGCUGUUCAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAAG--GUUAU- --.....((((....))))(((....(--(((((((.......(((.....(((((((.......)))))))..)))(((.......))).))))))))...))).--.....- ( -35.40, z-score = -0.39, R) >droEre2.scaffold_4929 6356355 110 + 26641161 --AUUUUAAGCGAACGCUGAGUAACUG--GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAGAAAAGAAAU --......(((....)))((((.....--)))).((..(((..((((....(((((((.......)))))))((((.(((.......)))))))..))))...)))..)).... ( -35.00, z-score = -0.34, R) >anoGam1.chr2R 51331534 98 - 62725911 ------------CUCUUCAACCACGUG--GCUCAAUGGUCUAGGGGUAUGAUUCUCGCUUUGGGUGCGAGAGGUCCCGGGUUCAAAUCCCGGUUGAGCCCGCGAUUUCUUUU-- ------------...........((((--(((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))).))))).........-- ( -35.00, z-score = -1.37, R) >consensus __AUUUUAAGCGCUCAUUGAUCAACUG__GCUCGUUGGUCUAGGGGUAUGAUUUCCGCUUAGGGUGCGGGAGGUCCCGGGUUCAAAUCCCGGACGAGCCCAAGAAAAAGUAAU_ .............................(((((((.......(((.....(((((((.......)))))))..)))(((.......))).)))))))................ (-30.01 = -28.97 + -1.04)
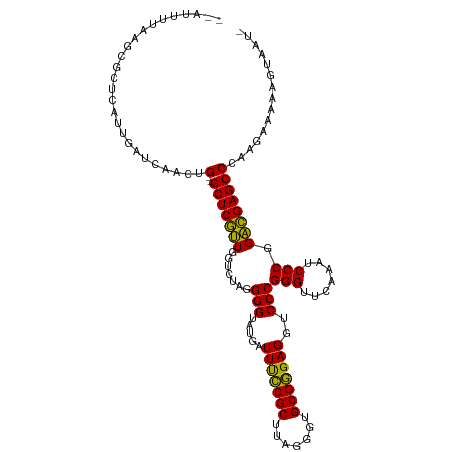
| Location | 14,598,834 – 14,598,944 |
|---|---|
| Length | 110 |
| Sequences | 11 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 79.05 |
| Shannon entropy | 0.47958 |
| G+C content | 0.53087 |
| Mean single sequence MFE | -31.47 |
| Consensus MFE | -23.27 |
| Energy contribution | -22.38 |
| Covariance contribution | -0.89 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.95 |
| SVM RNA-class probability | 0.976371 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14598834 110 - 23011544 AACUACUCCAUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGGUAAACAAGCGCUAAAAAU-- .......(((.((..((((((((((((.......)))))......((((.......))))...................))))))--))).)))..................-- ( -30.00, z-score = -1.10, R) >droVir3.scaffold_13047 2079034 107 + 19223366 -------AAUAUAAGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGCCGCACAUGAUCAAAGCAAGUUUUAAACAU -------........((((((((((((.......)))))((..((((((.......)))))).))..............)))))))((...........))............. ( -30.40, z-score = -3.21, R) >droWil1.scaffold_180764 2954693 109 - 3949147 -UUCAACUUCACUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGCCACAUUAAUGGCCAAAAG-- -...........(((((((((((((((.......)))))......((((.......))))...................))))))--)....((((......)))))))...-- ( -32.50, z-score = -2.10, R) >droAna3.scaffold_12916 14958224 109 + 16180835 -GUUAUUUUCACUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGUUCGACGAGCGACAGAAAC-- -.....((((.....((((((((((((.......)))))......((((.......))))...................))))))--).(((((((...))))))).)))).-- ( -36.10, z-score = -2.53, R) >dp4.chr4_group1 2039688 99 + 5278887 -----------CUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGACGUAUGGGCGGUUAAAAU-- -----------....((((((((((((.......)))))......((((.......))))...................))))))--)..(((((((....)).)))))...-- ( -29.80, z-score = 0.10, R) >droPer1.super_5 1983253 109 - 6813705 -AGUUAUUUUACUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGACGUAUGGGCGGUUAAAAU-- -....((((((....((((((((((((.......)))))......((((.......))))...................))))))--)......(((....)))..))))))-- ( -29.90, z-score = 0.40, R) >droSim1.chr2L 14350385 110 - 22036055 AACUACUCUUUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGUUCAUUGCGCGCUUAAAAU-- ............((((((.((((((((.......)))))......((((.......))))..............((.((((....--..)))).))...))).))))))...-- ( -28.20, z-score = -0.52, R) >droSec1.super_3 872538 110 - 7220098 AACUACUUCUUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGCUCACUGCGUGCUUAAAAU-- ...............((((((((((((.......)))))......((((.......))))...................))))))--)((((((.....))).)))......-- ( -30.20, z-score = -1.23, R) >droYak2.chr2L 14713281 107 - 22324452 -AUAAC--CUUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUGAACAGCGACUGCUAAUAAU-- -.....--.......((((((((((((.......)))))......((((.......))))...................))))))--)(((((.....))))).........-- ( -31.00, z-score = -1.93, R) >droEre2.scaffold_4929 6356355 110 - 26641161 AUUUCUUUUCUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC--CAGUUACUCAGCGUUCGCUUAAAAU-- ...............((((((((((((.......)))))......((((.......))))...................))))))--)........(((....)))......-- ( -29.60, z-score = -1.07, R) >anoGam1.chr2R 51331534 98 + 62725911 --AAAAGAAAUCGCGGGCUCAACCGGGAUUUGAACCCGGGACCUCUCGCACCCAAAGCGAGAAUCAUACCCCUAGACCAUUGAGC--CACGUGGUUGAAGAG------------ --......(((((((((((((((((((.......)))))((..((((((.......)))))).))..............))))))--).)))))))......------------ ( -38.50, z-score = -4.50, R) >consensus _AUUACUUUUUCUUGGGCUCGUCCGGGAUUUGAACCCGGGACCUCCCGCACCCUAAGCGGAAAUCAUACCCCUAGACCAACGAGC__CAGUUGAUCAAUGAGCGCUUAAAAU__ ................(((((((((((.......)))))......((((.......))))...................))))))............................. (-23.27 = -22.38 + -0.89)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:59 2011