| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,581,886 – 14,581,981 |
| Length | 95 |
| Max. P | 0.958942 |

| Location | 14,581,886 – 14,581,981 |
|---|---|
| Length | 95 |
| Sequences | 7 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 66.55 |
| Shannon entropy | 0.63138 |
| G+C content | 0.53926 |
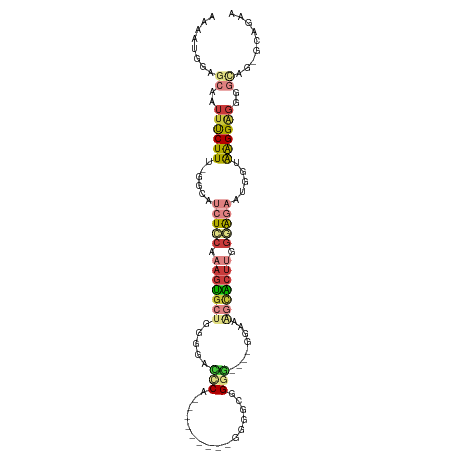
| Mean single sequence MFE | -31.77 |
| Consensus MFE | -11.19 |
| Energy contribution | -12.75 |
| Covariance contribution | 1.56 |
| Combinations/Pair | 1.61 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.35 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.66 |
| SVM RNA-class probability | 0.958942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
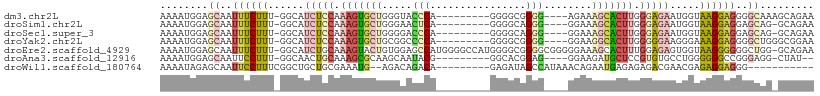
>dm3.chr2L 14581886 95 + 23011544 AAAAUGGAGCAAUUUCUUU-GGCAUCUCCAAAGUGCUGGGUACCCA---------GGGGCGGGG----AGAAAGCACUUGGGAGAAUGGUAAGGAGGGGCAAAGCAGAA ........((...((((((-(.(((((((.(((((((.....(((.---------.....))).----....))))))).)))).))).)))))))..))......... ( -35.00, z-score = -3.46, R) >droSim1.chr2L 14332015 94 + 22036055 AAAAUGGAGCAAUUUCUUU-GGCAUCUCCAAAGUGCUGGGAACUCA---------GGGGCAGGG----GGAAAGCACUUGGGAGAAUGGUAAGGAGGAGCAG-GCAGAA ........((..(((((((-(.(((((((.(((((((.....(((.---------......)))----....))))))).)))).))).)))))))).))..-...... ( -30.30, z-score = -2.69, R) >droSec1.super_3 855121 94 + 7220098 AAAAUGGAGCAAUUUCUUU-GGCAUCUCCAAAGUGCUGGGGACCCA---------GGGGCAGGG----GGAAAGCACUUGGGAGAAUGGUAAGGAGGAGCAG-GCAGAA ........((..(((((((-(.(((((((.(((((((.....(((.---------......)))----....))))))).)))).))).)))))))).))..-...... ( -33.30, z-score = -2.85, R) >droYak2.chr2L 14693569 95 + 22324452 AAAAUGGAGCAAUUUCUUU-GGCAUCUCCAAAGUGCUGCGGCCCCA---------GGGGCGGGG----GGAAGGCACUUGGGGGAAGGGAAAGGAGGGGCUGGGCGGAA .......(((..(((((((-..(.(((((.(((((((....((((.---------.......))----))..))))))).))))).)..)))))))..)))........ ( -36.60, z-score = -1.64, R) >droEre2.scaffold_4929 6336567 107 + 26641161 AAAAUGGAGCAAUUUCUUU-GGCAUCUGCAAAGUACUGUGGAGCCAUGGGGCCAUGGGGCGGGGCGGGGGAAAGCACUUUGGAGAGUGGUAAGGGGGGCUGG-GCAGAA ........((..(..((((-(.(((((.((((((.((((....(((((....))))).)))).((........)))))))).))).)).)))))..).....-)).... ( -32.60, z-score = -1.08, R) >droAna3.scaffold_12916 14943869 92 - 16180835 AAAAUGGAGCAAUUCCUUU-GGCAACUGCAAAGCGCAAGCAAUACG---------GGCACGGAG----GGAAGAUGCUCCGUGUGCCUGGGGGGCCGGGAGG-CUAU-- ..............(((..-(((..(((....((....))....))---------)((((((((----........)))))))))))..)))((((....))-))..-- ( -37.30, z-score = -2.32, R) >droWil1.scaffold_180764 1018704 87 - 3949147 AAAAUAGAGCAAUUCCUUUCGGCUGCUGCGAAAUG--AGACAGACA---------GAGAUAGCCAUAAACAGAAUGAGAGAGACGAACGAGAGGAGGG----------- ............(((((((((.(((((((......--.).))).))---------).......(((.......)))...........)))))))))..----------- ( -17.30, z-score = -1.69, R) >consensus AAAAUGGAGCAAUUUCUUU_GGCAUCUCCAAAGUGCUGGGGACCCA_________GGGGCGGGG____GGAAAGCACUUGGGAGAAUGGUAAGGAGGGGCAG_GCAGAA ........((..((((((......(((((.(((((((.....(((................)))........))))))).))))).....))))))..))......... (-11.19 = -12.75 + 1.56)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:54 2011