
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,310,146 – 1,310,287 |
| Length | 141 |
| Max. P | 0.836485 |

| Location | 1,310,146 – 1,310,287 |
|---|---|
| Length | 141 |
| Sequences | 7 |
| Columns | 160 |
| Reading direction | reverse |
| Mean pairwise identity | 66.29 |
| Shannon entropy | 0.65051 |
| G+C content | 0.56435 |
| Mean single sequence MFE | -55.29 |
| Consensus MFE | -20.16 |
| Energy contribution | -23.46 |
| Covariance contribution | 3.30 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.36 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.86 |
| SVM RNA-class probability | 0.836485 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1310146 141 - 23011544 ---CUGCCCCAAUCCCAUCCUGGGGCUUCGUUGA--GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUCCUAGU-------CUGGAUGCUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCCCGAGUGUGUGCG--AGGGGGUGAG----- ---..(((((..(((((..((.((((...(((((--.....)))))...((.((((((((((((((((........).)))))((-------(.(((.((....))))).))).)))))))))).)).)))).))..)).).)--).)))))...----- ( -58.70, z-score = -1.63, R) >droSim1.chr2L 1281549 129 - 22036055 ---CUGCCCCAAUCCCAUCCUGGAGCUCCGUUGA--GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUC-------------------CUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCCCGAGUGUGUGCA--AGGGGGUGAG----- ---..(((((.....((((((((((((......(--((..(.(((((((..((((((..(.......)..))))))...)-------------------))))))).))))))).))))))))((((..(......)..))))--..)))))...----- ( -52.00, z-score = -1.61, R) >droSec1.super_14 1259701 141 - 2068291 ---CUGCCCCAAUCCCAUCCUGGAGCUCCGUUGA--GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUCCUAGU-------CUGGAUGCUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCCCGAGUGUGUGCA--AGGGGGUGAG----- ---..(((((.....((((((((((((......(--((..(.((((((.....(((((.(((((((((........).)))))))-------).)))))))))))).))))))).))))))))((((..(......)..))))--..)))))...----- ( -56.80, z-score = -1.55, R) >droEre2.scaffold_4929 1348819 143 - 26641161 ---CUGCCCCACUCCCAUUCUAAACCUCCGUCGA--GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUCCUAGU-------CUGGAUGCUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCGGGAGGGGGGAGGGUGGGGGAUGAG----- ---((.((((.(((((......((((((....))--))))....(((..((.((((((((((((((((........).)))))((-------(.(((.((....))))).))).)))))))))).)))))))))))))).)).............----- ( -62.80, z-score = -1.45, R) >droYak2.chr2L 1286051 153 - 22324452 ---CUGCACCAAUCCCAUUCUAAACCUCCGUUGA--GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUCCUAGUCUGGAUGCUGGAUGCUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCCCGAGUGUGUGCG--GGGUGGCUGGUUGAG ---......((((((((((((.((((((....))--))))..(((((..(((.(((((.(((((((((........).)))))))).))))))))..)))))....((((((...)))))).(..((..(....)..))..))--))))))..))))).. ( -56.40, z-score = -0.67, R) >droAna3.scaffold_12916 4773603 131 - 16180835 ---------CACCUGCAAGCUUCGGCUCCGCUGA--CGUUGUCAGCAGGUAUAUGUCCUGGCUAGGACUCGACAUUGUCCUUAGU-------CUGGAUGCUGAGGCCCAGUCCUGU----GUGCGACGGCUCCAAGACGUGUGUGAGUGUGUG------- ---------(((..(((.((((((((.(((((((--(...)))))).)).....((((.(((((((((........)))).))))-------).))))))))))))...(((.((.----(.((....)).))).)))...)))..)))....------- ( -50.90, z-score = -1.52, R) >droGri2.scaffold_15252 6789268 145 + 17193109 CUCCUAUUCUCUGUGUAUACUGGUCUCAUGGUCAUGUGUAGACAGCUAGCAAUAAUUCUGAUACUGCCUCAGAGCUAACUCUGCU-------UCUUCUCCUGCAGCU-GUUGUUGCCGGGAAGAGGUAGCUCUGCUGUAUUCCAAACAUUGUG------- ..........((((.(((((((..(....)..)).))))).))))...(((((......(((((.((...((((((......(((-------(((((.((.(((((.-...))))).)))))))))))))))))).)))))......))))).------- ( -49.40, z-score = -3.45, R) >consensus ___CUGCCCCAAUCCCAUCCUGGAGCUCCGUUGA__GGUUGUCAGCAGGUAGAUGUCCUGGCUAGGACUCGACAUUGUUCCUAGU_______CUGGAUGCUGUUGCUCCUGGCUGCCAGGAUGUGUGUGCCCGAGUGUGUGCG__AGGGGGUGAG_____ ......((((...................(((((.......))))).((((.(((((((((((((((..(((((..((..((((........))))..))))))).)))))...))))))))..)).))))...............)))).......... (-20.16 = -23.46 + 3.30)

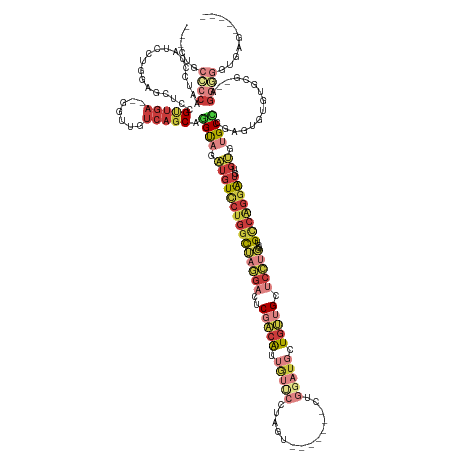
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:19 2011