
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,532,168 – 14,532,235 |
| Length | 67 |
| Max. P | 0.734283 |

| Location | 14,532,168 – 14,532,235 |
|---|---|
| Length | 67 |
| Sequences | 8 |
| Columns | 80 |
| Reading direction | forward |
| Mean pairwise identity | 75.28 |
| Shannon entropy | 0.44666 |
| G+C content | 0.53854 |
| Mean single sequence MFE | -20.20 |
| Consensus MFE | -8.71 |
| Energy contribution | -8.97 |
| Covariance contribution | 0.27 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.11 |
| Structure conservation index | 0.43 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.54 |
| SVM RNA-class probability | 0.734283 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
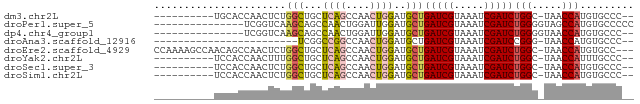
>dm3.chr2L 14532168 67 + 23011544 ----------UGCACCAACUCUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC-UAACCAUGUGCCC-- ----------.((((......(((((....)))))..(((..((((((((.....))))).)))-...))).))))..-- ( -22.90, z-score = -3.47, R) >droPer1.super_5 1899802 65 + 6813705 ---------------UCGGUCAAGCAGCCAACUGGAUUGGAUGCUGAUCGUAAAUCGAUCUGGGGUAGCCAUGUGCCCCC ---------------.((((((.(((.((((.....)))).)))))))))...........((((((......)))))). ( -23.00, z-score = -1.57, R) >dp4.chr4_group1 1956600 63 - 5278887 ---------------UCGGUCAAGCAGCCAACUGGAUUGGAUGCUGAUCGUAAAUCGAUCUGGGGUAACCAUGUGCCC-- ---------------.((((((.(((.((((.....)))).)))))))))............(((((......)))))-- ( -19.00, z-score = -0.97, R) >droAna3.scaffold_12916 14894058 53 - 16180835 ------------------------UCGGCCGGCCAACUGGAUGCUGAUCGUAAAUCGAUCCGGG-UAACCAUGUGCCC-- ------------------------......(((((..(((.(((((((((.....)))))..))-)).))))).))).-- ( -16.40, z-score = -0.73, R) >droEre2.scaffold_4929 6288580 76 + 26641161 CCAAAAGCCAACAGCCAACUCUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC-UAACCAUGUGCC--- .....(((((.((((((....)))))).((((....)))).....(((((.....)))))))))-)...........--- ( -22.60, z-score = -2.26, R) >droYak2.chr2L 14642539 67 + 22324452 ----------UCCACCAACUUUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC-UAACCAUUUGCCC-- ----------..........((((((....)))))).(((..((((((((.....))))).)))-...))).......-- ( -18.90, z-score = -2.75, R) >droSec1.super_3 795747 67 + 7220098 ----------UCCACCAACUCUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC-UAACCAUGUGCCC-- ----------..(((......(((((....)))))..(((..((((((((.....))))).)))-...))).)))...-- ( -19.40, z-score = -2.56, R) >droSim1.chr2L 14280721 67 + 22036055 ----------UCCACCAACUCUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC-UAACCAUGUGCCC-- ----------..(((......(((((....)))))..(((..((((((((.....))))).)))-...))).)))...-- ( -19.40, z-score = -2.56, R) >consensus __________UCCACCAACUCUGGCUGCUCAGCCAACUGGAUGCUGAUCGUAAAUCGAUCUGGC_UAACCAUGUGCCC__ ......................(((...((((....))))..)))(((((.....)))))(((.....)))......... ( -8.71 = -8.97 + 0.27)
| Location | 14,532,168 – 14,532,235 |
|---|---|
| Length | 67 |
| Sequences | 8 |
| Columns | 80 |
| Reading direction | reverse |
| Mean pairwise identity | 75.28 |
| Shannon entropy | 0.44666 |
| G+C content | 0.53854 |
| Mean single sequence MFE | -21.57 |
| Consensus MFE | -11.10 |
| Energy contribution | -11.21 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.55 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.555992 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
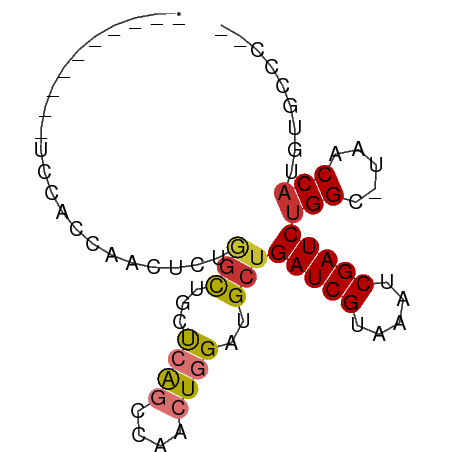
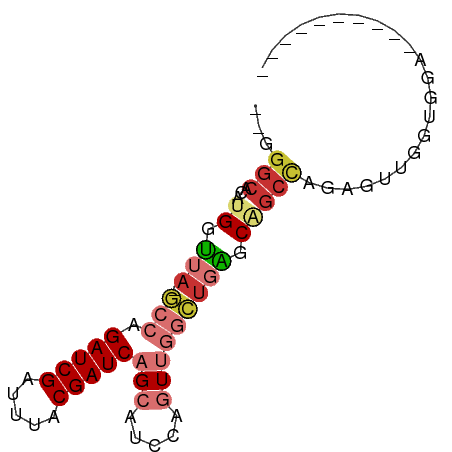
>dm3.chr2L 14532168 67 - 23011544 --GGGCACAUGGUUA-GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAGAGUUGGUGCA---------- --.(((.........-))).(((((.....))))).(((.(((.((((((....))))))...)))))).---------- ( -24.50, z-score = -2.38, R) >droPer1.super_5 1899802 65 - 6813705 GGGGGCACAUGGCUACCCCAGAUCGAUUUACGAUCAGCAUCCAAUCCAGUUGGCUGCUUGACCGA--------------- ((((((.....))..)))).(((((.....)))))((((.(((((...))))).)))).......--------------- ( -21.60, z-score = -1.56, R) >dp4.chr4_group1 1956600 63 + 5278887 --GGGCACAUGGUUACCCCAGAUCGAUUUACGAUCAGCAUCCAAUCCAGUUGGCUGCUUGACCGA--------------- --.......((((((.....(((((.....)))))((((.(((((...))))).)))))))))).--------------- ( -19.00, z-score = -1.78, R) >droAna3.scaffold_12916 14894058 53 + 16180835 --GGGCACAUGGUUA-CCCGGAUCGAUUUACGAUCAGCAUCCAGUUGGCCGGCCGA------------------------ --.(((....(((((-.(.(((((((((...)))).).)))).).))))).)))..------------------------ ( -14.60, z-score = -0.00, R) >droEre2.scaffold_4929 6288580 76 - 26641161 ---GGCACAUGGUUA-GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAGAGUUGGCUGUUGGCUUUUGG ---.((...(((...-.)))(((((.....))))).))..((((..((((.(((((((......))))))))))).)))) ( -29.30, z-score = -2.59, R) >droYak2.chr2L 14642539 67 - 22324452 --GGGCAAAUGGUUA-GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAAAGUUGGUGGA---------- --.(((...((.(((-(((.(((((.....)))))(((.....))))))))).)))))............---------- ( -21.20, z-score = -1.58, R) >droSec1.super_3 795747 67 - 7220098 --GGGCACAUGGUUA-GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAGAGUUGGUGGA---------- --.(((...((.(((-(((.(((((.....)))))(((.....))))))))).)))))............---------- ( -21.20, z-score = -1.26, R) >droSim1.chr2L 14280721 67 - 22036055 --GGGCACAUGGUUA-GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAGAGUUGGUGGA---------- --.(((...((.(((-(((.(((((.....)))))(((.....))))))))).)))))............---------- ( -21.20, z-score = -1.26, R) >consensus __GGGCACAUGGUUA_GCCAGAUCGAUUUACGAUCAGCAUCCAGUUGGCUGAGCAGCCAGAGUUGGUGGA__________ ...(((...(((.....)))(((((.....)))))....................)))...................... (-11.10 = -11.21 + 0.11)

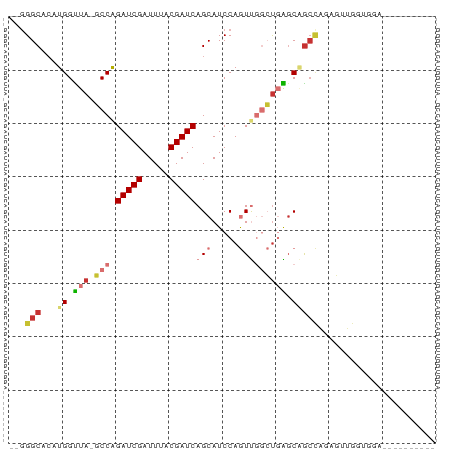
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:46 2011