
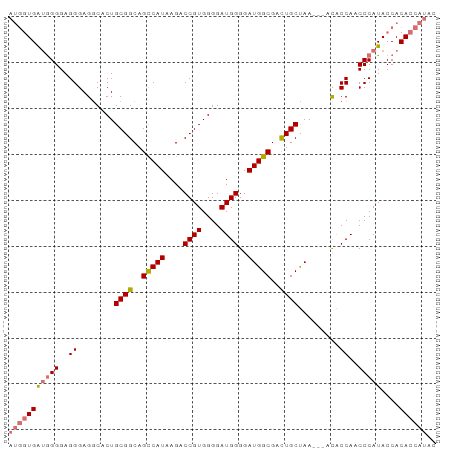
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,531,279 – 14,531,440 |
| Length | 161 |
| Max. P | 0.877012 |

| Location | 14,531,279 – 14,531,369 |
|---|---|
| Length | 90 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 91.26 |
| Shannon entropy | 0.13957 |
| G+C content | 0.58721 |
| Mean single sequence MFE | -34.59 |
| Consensus MFE | -28.69 |
| Energy contribution | -29.63 |
| Covariance contribution | 0.94 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.15 |
| Structure conservation index | 0.83 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.877012 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14531279 90 - 23011544 AUGGUGAUGGGGAGGGAGACACUGCGGCAGCCAUAAGACCGUGGGGAUGGGGAUGGCGACUGCUAA---ACACCAACCCAUACCACACCAUAC (((((((((((..((........((((..(((((....((((....))))..)))))..))))...---...))..)))))....)))))).. ( -35.14, z-score = -2.97, R) >droSim1.chr2L 14279828 90 - 22036055 AUGGUGAUGGGGAGGGAGGCACUGCGGCAGCCAUAAGACCGUGGGGAUGGGGAUGGCGACUGCUAA---ACACCAACCCAUACCACACCAUAC ((((((.(((...(((.((....((((..(((((....((((....))))..)))))..))))...---...))..)))...))))))))).. ( -35.40, z-score = -2.50, R) >droSec1.super_3 794858 90 - 7220098 AUGGUGAUGGGGAGGGAGGCACUGCGGCAGUCAUAAGACCGUGGGGAUGGGGAUGGCGACUGCUAA---ACACCAACCCAUACCACACCAUAC ((((((.(((...(((.((....((((..(((((....((((....))))..)))))..))))...---...))..)))...))))))))).. ( -32.70, z-score = -1.77, R) >droYak2.chr2L 14641631 91 - 22324452 --AUUGGGAGGGAGGGAGGCACUGCGGCAGCCAUAAGACCGUGGGGAUGGGGAUGGCGAUUGCCAACCCAUGCCAACCCAUACCACACCAUAC --..((((.((..(((.(((.(..((((........).)))..)..(((((..((((....)))).))))))))..)))...)).).)))... ( -35.10, z-score = -1.38, R) >consensus AUGGUGAUGGGGAGGGAGGCACUGCGGCAGCCAUAAGACCGUGGGGAUGGGGAUGGCGACUGCUAA___ACACCAACCCAUACCACACCAUAC (((((((((((..((........((((..(((((....((((....))))..)))))..)))).........))..)))))....)))))).. (-28.69 = -29.63 + 0.94)


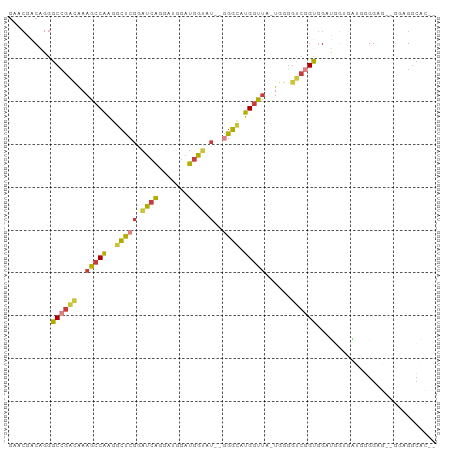
| Location | 14,531,347 – 14,531,440 |
|---|---|
| Length | 93 |
| Sequences | 7 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 71.68 |
| Shannon entropy | 0.56463 |
| G+C content | 0.62289 |
| Mean single sequence MFE | -28.77 |
| Consensus MFE | -13.83 |
| Energy contribution | -13.84 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.58 |
| Mean z-score | -1.11 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.572834 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14531347 93 - 23011544 GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU--GGGCAUGGUUA-UGGGGUCGGUGGAUGGUGAUGGGGAG--GGAGACAC-- ...(.((..((((..((.(((((..(((((.((((.......)))).)--)))).))))).-)).)))).)).)..............--(....)..-- ( -25.40, z-score = -1.77, R) >droSim1.chr2L 14279896 93 - 22036055 GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU--GGGCAUGGUUA-UGGGGUCGGUGGAUGGUGAUGGGGAG--GGAGGCAC-- ...(.((..((((..((.(((((..(((((.((((.......)))).)--)))).))))).-)).)))).)).)..............--........-- ( -23.90, z-score = -0.99, R) >droSec1.super_3 794926 93 - 7220098 GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU--GGGCAUGGUUA-UGGGGUCGGUGGAUGGUGAUGGGGAG--GGAGGCAC-- ...(.((..((((..((.(((((..(((((.((((.......)))).)--)))).))))).-)).)))).)).)..............--........-- ( -23.90, z-score = -0.99, R) >droYak2.chr2L 14641702 91 - 22324452 GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU--GGGCAUGGUUA-UGGGGUCGGUGGAUUGGGA--GGGAG--GGAGGCAC-- ......(((.((((((..(((((..(((((.((((.......)))).)--)))).))))).-....))))))...)))...--.....--........-- ( -24.00, z-score = -1.60, R) >droEre2.scaffold_4929 6287742 86 - 26641161 GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU--GGGCAUGGUUA-UGGGGUCGGUGGAU-------GGGGA--GGAGGCAC-- ...(..((..((((((..(((((..(((((.((((.......)))).)--)))).))))).-....))))))...)-------)..).--........-- ( -24.50, z-score = -1.85, R) >droAna3.scaffold_12916 14893156 99 + 16180835 GAACGGCGUGGCCGACAAAGCCAAGGCUCGGAUCAGGACGGAUGGAGUCAAGGGAUGGUUG-GUGAAGGAGUGGCCCGGGGCUCGGAGCUGGAGGGGCAC ........(.((((((....((..(((((.(.((......))).)))))...))...))))-)).)....(((.(((..(((.....)))...))).))) ( -31.60, z-score = 0.18, R) >droMoj3.scaffold_6500 16484064 93 + 32352404 ----GACAUGGCAGCUGCCGGCCCGGACCAGUCGGCCCCAGUUGGGCCCCAGUUGGGGCUGUUGGGGGCAGCUGAGGCAACUGAGGCAACUGGGGCA--- ----(((..(((....)))((......)).))).((((((((((((((((....))))))...........((.((....)).)).)))))))))).--- ( -48.10, z-score = -0.73, R) >consensus GAACGACAGGGCCGACAAAGCCAAGGCUCGGAUCAGGAUGGAUGGUAU__GGGCAUGGUUA_UGGGGUCGGUGGAUGGUGAUGGGGAG__GGAGGCAC__ ..........((((((..(((((..((((..((((.......))))....)))).)))))......))))))............................ (-13.83 = -13.84 + 0.01)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:45 2011