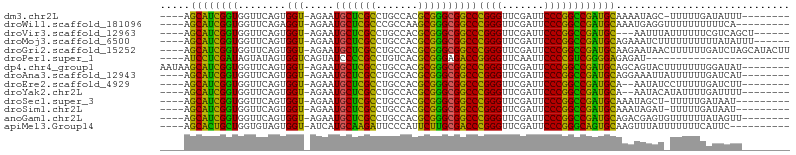
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,530,526 – 14,530,617 |
| Length | 91 |
| Max. P | 0.994108 |

| Location | 14,530,526 – 14,530,617 |
|---|---|
| Length | 91 |
| Sequences | 14 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 79.06 |
| Shannon entropy | 0.47174 |
| G+C content | 0.55919 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -29.71 |
| Energy contribution | -29.32 |
| Covariance contribution | -0.38 |
| Combinations/Pair | 1.40 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.994108 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
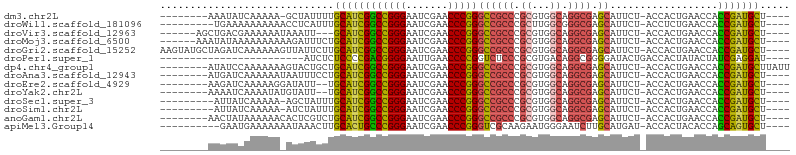
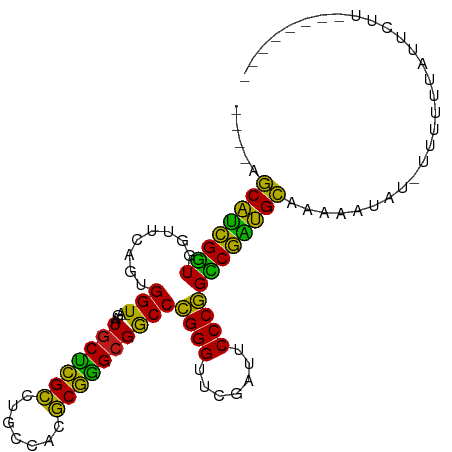
>dm3.chr2L 14530526 91 + 23011544 --------AAAUAUCAAAAA-GCUAUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- --------............-........((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -1.85, R) >droWil1.scaffold_181096 4526278 91 + 12416693 ---------UGAAAAAAAAAACCUCAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCU-ACCUCUGAACCACCGAUGCU---- ---------....................((((((((((((.......)))))(((((((((...))))))).)).....-............))))))).---- ( -38.30, z-score = -3.95, R) >droVir3.scaffold_12963 205121 91 - 20206255 ------AGCUGACGAAAAAAUAAAUU---GCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- ------....................---((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -1.72, R) >droMoj3.scaffold_6500 23190625 94 + 32352404 ------AAAUAUAAAAAAAAAAGAUUUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- ------.......................((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -2.28, R) >droGri2.scaffold_15252 5723939 100 - 17193109 AAGUAUGCUAGAUCAAAAAAGUUAUUCUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- .............................((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -0.96, R) >droPer1.super_1 8485218 77 + 10282868 ------------------------AUCUCUCCCCGACGGGGAAUUGAACCCCGGUCUCCCGCGUGACAGGCGGGGAUACUGACCACUAUACUAUCGAGGAU---- ------------------------.....(((.(((.((((.......))))((((((((((.......)))))).....)))).........))).))).---- ( -28.20, z-score = -1.19, R) >dp4.chr4_group1 4155954 96 - 5278887 --------AUAUCCAAAAAAAGUACUGCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCUUAUU --------.....................((((((((((((.......)))))((((((.((...)).)))).)).....-............)))))))..... ( -32.20, z-score = -1.49, R) >droAna3.scaffold_12943 3040220 92 + 5039921 --------AUGAUCAAAAAAUAAUUUCCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- --------.....................((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -1.84, R) >droEre2.scaffold_4929 6286843 90 + 26641161 --------AAGAUCAAAAAGGAUAUU--UGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- --------...(((......)))...--.((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.30, z-score = -1.55, R) >droYak2.chr2L 14640779 90 + 22324452 --------AAAAUCAAAAUAUGUAUU--UGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- --------..................--.((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -1.88, R) >droSec1.super_3 794187 90 + 7220098 ---------AUUAUCAAAAA-AGCUAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- ---------...........-........((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -1.84, R) >droSim1.chr2L 14279161 90 + 22036055 ---------AUUAUCAAAAA-AUCUAUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- ---------...........-........((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -2.12, R) >anoGam1.chr2L 14910695 92 + 48795086 --------AACUAUAAAAAACACUCGUCUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCACCGAUGCU---- --------.....................((((((((((((.......)))))((((((.((...)).)))).)).....-............))))))).---- ( -32.20, z-score = -2.04, R) >apiMel3.Group14 6137489 90 - 8318479 ----------GAAUGAAAAAAAUAAACUUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAU-ACCACUACACCAGCAGUGCU---- ----------...................((((((((((((.......)))))...((((((.......)))))).....-............))))))).---- ( -29.30, z-score = -3.73, R) >consensus ________AAGAACAAAAAA_AUAUUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU_ACCACUGAACCACCGAUGCU____ .............................((((((((((((.......)))))((((((.((...)).)))).))..................)))))))..... (-29.71 = -29.32 + -0.38)
| Location | 14,530,526 – 14,530,617 |
|---|---|
| Length | 91 |
| Sequences | 14 |
| Columns | 105 |
| Reading direction | reverse |
| Mean pairwise identity | 79.06 |
| Shannon entropy | 0.47174 |
| G+C content | 0.55919 |
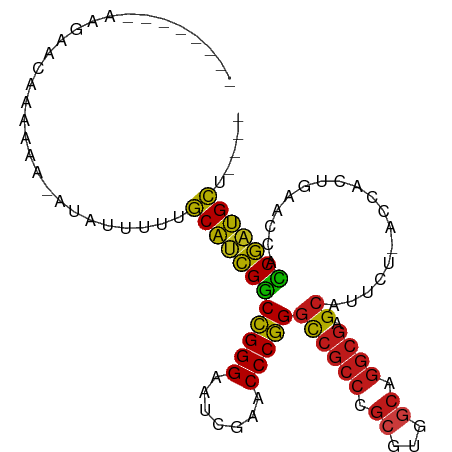
| Mean single sequence MFE | -35.54 |
| Consensus MFE | -33.74 |
| Energy contribution | -33.11 |
| Covariance contribution | -0.62 |
| Combinations/Pair | 1.52 |
| Mean z-score | -1.70 |
| Structure conservation index | 0.95 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.31 |
| SVM RNA-class probability | 0.988161 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14530526 91 - 23011544 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAUAGC-UUUUUGAUAUUU-------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))........-............-------- ( -35.90, z-score = -1.51, R) >droWil1.scaffold_181096 4526278 91 - 12416693 ----AGCAUCGGUGGUUCAGAGGU-AGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUGAGGUUUUUUUUUUCA--------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))...(((((.......)))))--------- ( -38.70, z-score = -2.23, R) >droVir3.scaffold_12963 205121 91 + 20206255 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGC---AAUUUAUUUUUUCGUCAGCU------ ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))---....................------ ( -35.90, z-score = -1.43, R) >droMoj3.scaffold_6500 23190625 94 - 32352404 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGAAAUCUUUUUUUUUUAUAUUU------ ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).......................------ ( -35.90, z-score = -1.97, R) >droGri2.scaffold_15252 5723939 100 + 17193109 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAGAAUAACUUUUUUGAUCUAGCAUACUU ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))............................. ( -35.90, z-score = -1.05, R) >droPer1.super_1 8485218 77 - 10282868 ----AUCCUCGAUAGUAUAGUGGUCAGUAUCCCCGCCUGUCACGCGGGAGACCGGGGUUCAAUUCCCCGUCGGGGAGAGAU------------------------ ----.((((((((........((((......(((((.......))))).))))((((.......)))))))))))).....------------------------ ( -31.80, z-score = -1.83, R) >dp4.chr4_group1 4155954 96 + 5278887 AAUAAGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGCAGUACUUUUUUUGGAUAU-------- .....(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).....................-------- ( -35.90, z-score = -1.03, R) >droAna3.scaffold_12943 3040220 92 - 5039921 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGGAAAUUAUUUUUUGAUCAU-------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).....................-------- ( -35.90, z-score = -1.29, R) >droEre2.scaffold_4929 6286843 90 - 26641161 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCA--AAUAUCCUUUUUGAUCUU-------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).--..................-------- ( -35.90, z-score = -1.52, R) >droYak2.chr2L 14640779 90 - 22324452 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCA--AAUACAUAUUUUGAUUUU-------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).--..................-------- ( -35.90, z-score = -1.85, R) >droSec1.super_3 794187 90 - 7220098 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUAGCU-UUUUUGAUAAU--------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))........-...........--------- ( -35.90, z-score = -1.52, R) >droSim1.chr2L 14279161 90 - 22036055 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAUAGAU-UUUUUGAUAAU--------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))))........-...........--------- ( -35.90, z-score = -1.74, R) >anoGam1.chr2L 14910695 92 - 48795086 ----AGCAUCGGUGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAGACGAGUGUUUUUUAUAGUU-------- ----.(((((((..((((......-.))))((.(((((((...)))))))))(((((.......)))))))))))).....................-------- ( -35.90, z-score = -1.22, R) >apiMel3.Group14 6137489 90 + 8318479 ----AGCACUGCUGGUGUAGUGGU-AUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAAGUUUAUUUUUUUCAUUC---------- ----.((((((((((((......)-))))(((((((.......)))))))..(((((.......))))))))))))...................---------- ( -32.20, z-score = -3.68, R) >consensus ____AGCAUCGGUGGUUCAGUGGU_AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAAAUAU_UUUUUUAUUCUU________ .....(((((((..((((........))))((.(((((((...)))))))))(((((.......))))))))))))............................. (-33.74 = -33.11 + -0.62)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:43 2011