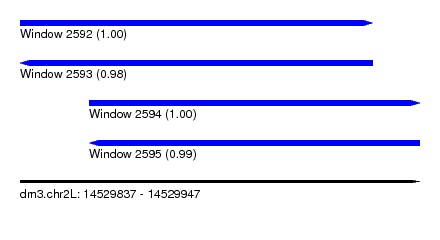
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,529,837 – 14,529,947 |
| Length | 110 |
| Max. P | 0.998616 |

| Location | 14,529,837 – 14,529,934 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 89.59 |
| Shannon entropy | 0.19686 |
| G+C content | 0.44988 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -28.45 |
| Energy contribution | -28.01 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.07 |
| Mean z-score | -3.25 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.05 |
| SVM RNA-class probability | 0.997178 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
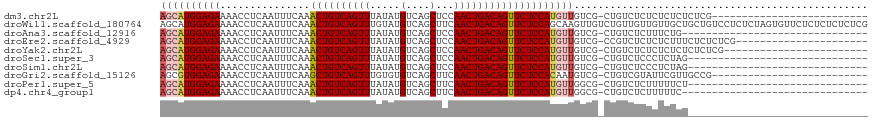
>dm3.chr2L 14529837 97 + 23011544 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUC---------------------- .............((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-)....---------------------- ( -30.16, z-score = -3.18, R) >droWil1.scaffold_180764 2851130 120 + 3949147 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUGUAUGUCAGCUUCAACUGACAGUUCUCCAGCAAGUUGUCUGUUGUUGUUGCUGCUGUCCUCUCUA (((((((((...))).)))))).((((((..............(((((((((((.((......))...)))))))))))...(((((.((...(.......)..)))))))...)))))) ( -33.80, z-score = -1.78, R) >droAna3.scaffold_12916 14892182 95 - 16180835 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUG------------------------ .............((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-)..------------------------ ( -30.16, z-score = -3.39, R) >droEre2.scaffold_4929 6286129 99 + 26641161 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CCGUCUC-------------------- ...((((......((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-)))))..-------------------- ( -32.46, z-score = -4.19, R) >droYak2.chr2L 14640058 99 + 22324452 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUC-------------------- ...((((.....)((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-).)))..-------------------- ( -31.36, z-score = -3.60, R) >droSec1.super_3 793470 95 + 7220098 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUG------------------------ .............((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-)..------------------------ ( -30.16, z-score = -3.39, R) >droSim1.chr2L 14278428 95 + 22036055 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUG------------------------ .............((.(.(((((((((((...............((((((((((.....(....)...))))))))))))))))))))).))-)..------------------------ ( -30.16, z-score = -3.39, R) >droGri2.scaffold_15126 878179 99 + 8399593 GUUGACGUGUUACGCUGUCAGCGUGGAGAAAACCUCAAUUUCAAGCUGUCAGUUUGUGUGUCAGCUUCAACUGACAGUUCUCCACAAUGUCG-CUGUCGU-------------------- (((((((((...))).))))))(((((((...............((((((((((...((....))...))))))))))))))))).......-.......-------------------- ( -32.76, z-score = -2.41, R) >droVir3.scaffold_12723 3152629 99 - 5802038 GUUGACGUGUUACGCUGUCAGCGUGGAGAAAACCUCAAUUUCAAGCUGUCAGUUUGUAUGUCAGCUCCAACUGACAGUUCUCCACAAUGUCG-UUGUCGU-------------------- (((((((((...))).))))))(((((((...............((((((((((.((......))...))))))))))))))))).......-.......-------------------- ( -31.96, z-score = -2.93, R) >droMoj3.scaffold_6500 16482965 99 - 32352404 GUUGACGUGUUACGCUGUCAGCGUGGAGAAAACCUCAAUUUCAAGCUGUCAGUUUGUAUGUCAGCUCCAACUGACAGUUCUCCACAAUGUCG-UUGUCGU-------------------- (((((((((...))).))))))(((((((...............((((((((((.((......))...))))))))))))))))).......-.......-------------------- ( -31.96, z-score = -2.93, R) >droPer1.super_5 1897589 95 + 6813705 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCG-CUG------------------------ .............((.(((((((((((((...............((((((((((.....(....)...))))))))))))))))))))))))-)..------------------------ ( -33.46, z-score = -3.93, R) >dp4.chr4_group1 1954341 95 - 5278887 GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCG-CUG------------------------ .............((.(((((((((((((...............((((((((((.....(....)...))))))))))))))))))))))))-)..------------------------ ( -33.46, z-score = -3.93, R) >consensus GUUGACGUGUUACGCUGUCAGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG_CUGUC______________________ (((((((((...))).))))))(((((((...............((((((((((.....(....)...)))))))))))))))))................................... (-28.45 = -28.01 + -0.44)

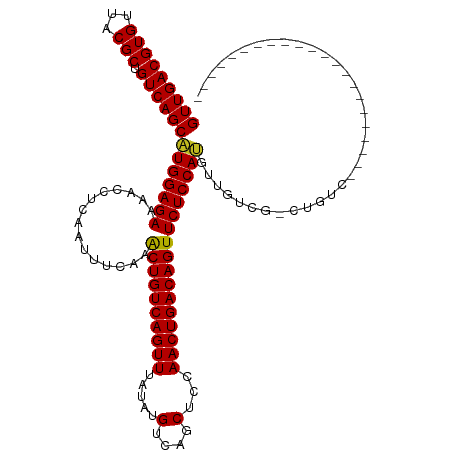
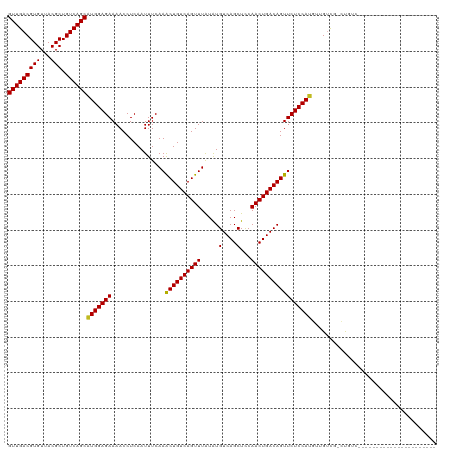
| Location | 14,529,837 – 14,529,934 |
|---|---|
| Length | 97 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 89.59 |
| Shannon entropy | 0.19686 |
| G+C content | 0.44988 |
| Mean single sequence MFE | -30.18 |
| Consensus MFE | -26.74 |
| Energy contribution | -26.08 |
| Covariance contribution | -0.66 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.42 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.984122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14529837 97 - 23011544 ----------------------GACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ----------------------(((.(-(..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))(.....))))... ( -30.04, z-score = -2.66, R) >droWil1.scaffold_180764 2851130 120 - 3949147 UAGAGAGGACAGCAGCAACAACAACAGACAACUUGCUGGAGAACUGUCAGUUGAAGCUGACAUACAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ..((((((((..((((((..............))))))..((((((((((((..............)))))))))))).........)))))))).((((.....))))........... ( -33.38, z-score = -1.69, R) >droAna3.scaffold_12916 14892182 95 + 16180835 ------------------------CAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ------------------------..(-(..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))............. ( -28.04, z-score = -2.12, R) >droEre2.scaffold_4929 6286129 99 - 26641161 --------------------GAGACGG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC --------------------..(((((-((.((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))....)))....))))... ( -31.44, z-score = -2.89, R) >droYak2.chr2L 14640058 99 - 22324452 --------------------GAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC --------------------..(((.(-(..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))(.....))))... ( -30.24, z-score = -2.56, R) >droSec1.super_3 793470 95 - 7220098 ------------------------CAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ------------------------..(-(..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))............. ( -28.04, z-score = -2.12, R) >droSim1.chr2L 14278428 95 - 22036055 ------------------------CAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ------------------------..(-(..((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))............. ( -28.04, z-score = -2.12, R) >droGri2.scaffold_15126 878179 99 - 8399593 --------------------ACGACAG-CGACAUUGUGGAGAACUGUCAGUUGAAGCUGACACACAAACUGACAGCUUGAAAUUGAGGUUUUCUCCACGCUGACAGCGUAACACGUCAAC --------------------..(((.(-(..((.(((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))).))...))(.....))))... ( -31.60, z-score = -2.28, R) >droVir3.scaffold_12723 3152629 99 + 5802038 --------------------ACGACAA-CGACAUUGUGGAGAACUGUCAGUUGGAGCUGACAUACAAACUGACAGCUUGAAAUUGAGGUUUUCUCCACGCUGACAGCGUAACACGUCAAC --------------------.......-.(((...((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))((.....)).......)))... ( -31.10, z-score = -2.57, R) >droMoj3.scaffold_6500 16482965 99 + 32352404 --------------------ACGACAA-CGACAUUGUGGAGAACUGUCAGUUGGAGCUGACAUACAAACUGACAGCUUGAAAUUGAGGUUUUCUCCACGCUGACAGCGUAACACGUCAAC --------------------.......-.(((...((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))((.....)).......)))... ( -31.10, z-score = -2.57, R) >droPer1.super_5 1897589 95 - 6813705 ------------------------CAG-CGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ------------------------..(-(((((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))))........... ( -29.54, z-score = -2.72, R) >dp4.chr4_group1 1954341 95 + 5278887 ------------------------CAG-CGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ------------------------..(-(((((.((((((((((((((((((..............))))))))))))((((.......)))))))))).))...))))........... ( -29.54, z-score = -2.72, R) >consensus ______________________GACAG_CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCUGACAGCGUAACACGUCAAC ...............................((.(((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))).))...(((.....))).... (-26.74 = -26.08 + -0.66)
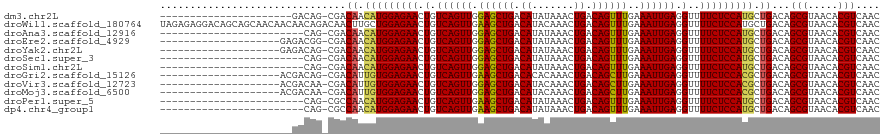
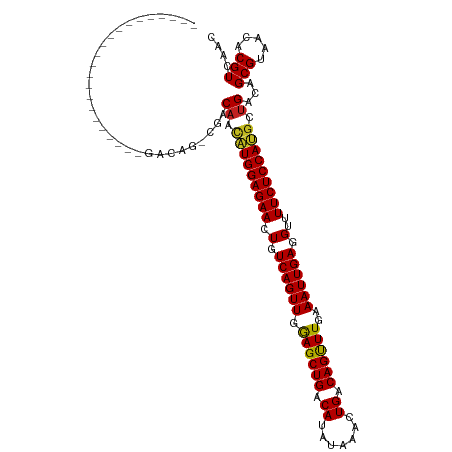
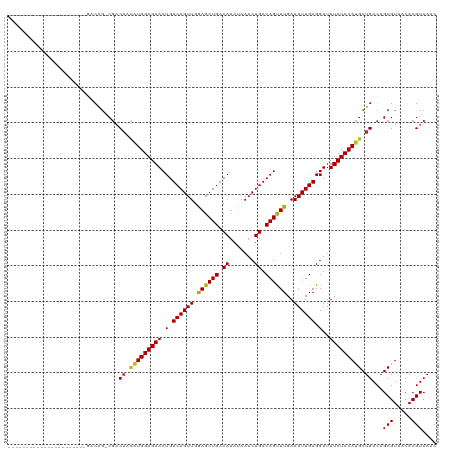
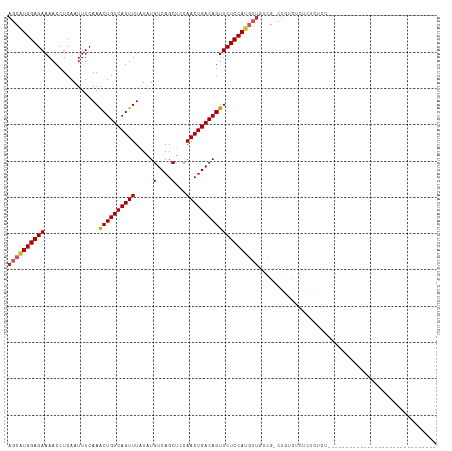
| Location | 14,529,856 – 14,529,947 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.31968 |
| G+C content | 0.43372 |
| Mean single sequence MFE | -24.66 |
| Consensus MFE | -22.63 |
| Energy contribution | -22.98 |
| Covariance contribution | 0.35 |
| Combinations/Pair | 1.10 |
| Mean z-score | -3.33 |
| Structure conservation index | 0.92 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 3.42 |
| SVM RNA-class probability | 0.998616 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14529856 91 + 23011544 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUCUCUCUCUCUCG-------------------------- ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-..................-------------------------- ( -24.06, z-score = -3.96, R) >droWil1.scaffold_180764 2851149 118 + 3949147 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUGUAUGUCAGCUUCAACUGACAGUUCUCCAGCAAGUUGUCUGUUGUUGUUGCUGCUGUCCUCUCUAGUGUUCUCUCUCUCUCG ((((((((((..............(((((((((((.((......))...)))))))))))...(((((.((...(.......)..)))))))...)))))).))))............ ( -27.80, z-score = -1.51, R) >droAna3.scaffold_12916 14892201 86 - 16180835 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUCUUUCUG------------------------------- ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-.............------------------------------- ( -24.06, z-score = -3.63, R) >droEre2.scaffold_4929 6286148 95 + 26641161 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CCGUCUCUCUCUUUCUCUCUCG---------------------- ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-......................---------------------- ( -24.06, z-score = -4.22, R) >droYak2.chr2L 14640077 93 + 22324452 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUCUCUCUCUCUCUCG------------------------ ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-....................------------------------ ( -24.06, z-score = -3.92, R) >droSec1.super_3 793489 87 + 7220098 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUCCCUCUAG------------------------------ ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-..............------------------------------ ( -24.06, z-score = -3.83, R) >droSim1.chr2L 14278447 87 + 22036055 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG-CUGUCUCCCUCUAG------------------------------ ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-..............------------------------------ ( -24.06, z-score = -3.83, R) >droGri2.scaffold_15126 878198 91 + 8399593 AGCGUGGAGAAAACCUCAAUUUCAAGCUGUCAGUUUGUGUGUCAGCUUCAACUGACAGUUCUCCACAAUGUCG-CUGUCGUAUUCGUUGCCG-------------------------- ...(((((((...............((((((((((...((....))...)))))))))))))))))......(-(...((....))..))..-------------------------- ( -26.36, z-score = -2.31, R) >droPer1.super_5 1897608 87 + 6813705 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCG-CUGUCUCUUUUUCU------------------------------ ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-..............------------------------------ ( -24.06, z-score = -3.03, R) >dp4.chr4_group1 1954360 86 - 5278887 AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUUCAACUGACAGUUCUCCAUGUUGGCG-CUGUCUCUUUUUC------------------------------- ((((((((((...............((((((((((.....(....)...))))))))))))))))))))....-.............------------------------------- ( -24.06, z-score = -3.09, R) >consensus AGCAUGGAGAAAACCUCAAUUUCAAACUGUCAGUUUAUAUGUCAGCUCCAACUGACAGUUCUCCAUGUUGUCG_CUGUCUCUCUCUCU______________________________ ((((((((((...............((((((((((.....(....)...))))))))))))))))))))................................................. (-22.63 = -22.98 + 0.35)
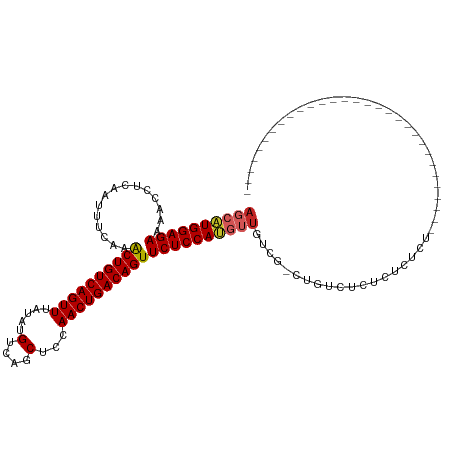
| Location | 14,529,856 – 14,529,947 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 82.50 |
| Shannon entropy | 0.31968 |
| G+C content | 0.43372 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -23.35 |
| Energy contribution | -22.98 |
| Covariance contribution | -0.37 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.18 |
| Structure conservation index | 0.91 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.39 |
| SVM RNA-class probability | 0.989942 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14529856 91 - 23011544 --------------------------CGAGAGAGAGAGAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU --------------------------..................-......((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -23.84, z-score = -1.72, R) >droWil1.scaffold_180764 2851149 118 - 3949147 CGAGAGAGAGAGAACACUAGAGAGGACAGCAGCAACAACAACAGACAACUUGCUGGAGAACUGUCAGUUGAAGCUGACAUACAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU .....(.((((((((.((...........((((((..............))))))..((((((((((((..............)))))))))))).......)))))))))))..... ( -31.88, z-score = -1.95, R) >droAna3.scaffold_12916 14892201 86 + 16180835 -------------------------------CAGAAAGAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU -------------------------------.............-......((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -23.84, z-score = -2.20, R) >droEre2.scaffold_4929 6286148 95 - 26641161 ----------------------CGAGAGAGAAAGAGAGAGACGG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ----------------------....................(.-...)..((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -24.14, z-score = -1.67, R) >droYak2.chr2L 14640077 93 - 22324452 ------------------------CGAGAGAGAGAGAGAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ------------------------....................-......((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -23.84, z-score = -1.58, R) >droSec1.super_3 793489 87 - 7220098 ------------------------------CUAGAGGGAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ------------------------------......(((((.((-(..(((......((((((((((((..............))))))))))))....)))..))).)))))..... ( -26.44, z-score = -2.61, R) >droSim1.chr2L 14278447 87 - 22036055 ------------------------------CUAGAGGGAGACAG-CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ------------------------------......(((((.((-(..(((......((((((((((((..............))))))))))))....)))..))).)))))..... ( -26.44, z-score = -2.61, R) >droGri2.scaffold_15126 878198 91 - 8399593 --------------------------CGGCAACGAAUACGACAG-CGACAUUGUGGAGAACUGUCAGUUGAAGCUGACACACAAACUGACAGCUUGAAAUUGAGGUUUUCUCCACGCU --------------------------((....))..........-.......((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))... ( -27.90, z-score = -2.59, R) >droPer1.super_5 1897608 87 - 6813705 ------------------------------AGAAAAAGAGACAG-CGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ------------------------------..............-......((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -23.84, z-score = -2.38, R) >dp4.chr4_group1 1954360 86 + 5278887 -------------------------------GAAAAAGAGACAG-CGCCAACAUGGAGAACUGUCAGUUGAAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU -------------------------------.............-......((((((((((((((((((..............))))))))))))((((.......)))))))))).. ( -23.84, z-score = -2.51, R) >consensus ______________________________AGAGAGAGAGACAG_CGACAACAUGGAGAACUGUCAGUUGGAGCUGACAUAUAAACUGACAGUUUGAAAUUGAGGUUUUCUCCAUGCU ...................................................(((((((((.(.((((((.((((((.((.......)).))))))..)))))).)..))))))))).. (-23.35 = -22.98 + -0.37)
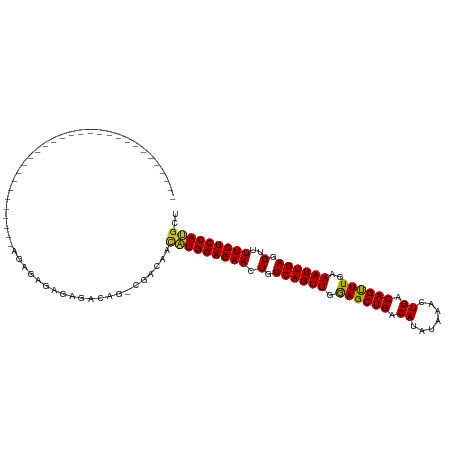
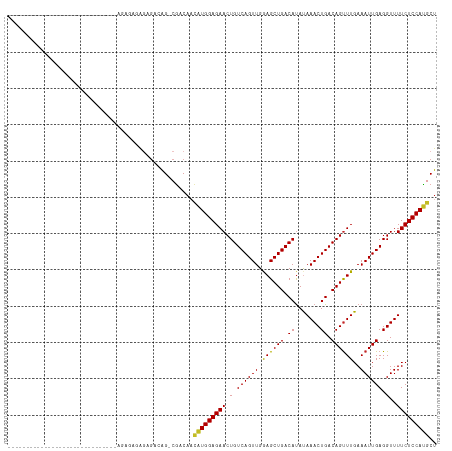
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:42 2011