
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,529,544 – 14,529,654 |
| Length | 110 |
| Max. P | 0.680894 |

| Location | 14,529,544 – 14,529,654 |
|---|---|
| Length | 110 |
| Sequences | 10 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 75.19 |
| Shannon entropy | 0.48986 |
| G+C content | 0.43484 |
| Mean single sequence MFE | -21.35 |
| Consensus MFE | -14.34 |
| Energy contribution | -14.62 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -0.88 |
| Structure conservation index | 0.67 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.680894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14529544 110 - 23011544 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC-ACAAAAGCCCAGGACGUUAUACGGAGGUAUGGCCAGAUGAAUGCAAA- ..((((...(((((......(((.((((((.....)))))).))).....))))).))))...-......((.((....(((((((....)))))))....))...))...- ( -24.10, z-score = -0.92, R) >droAna3.scaffold_12916 14891866 110 + 16180835 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC-AC-AAAGCACAGAACGGCGUACUCAGAUACAGCCAGAUGAAUGCAAAU ..((((...(((((......(((.((((((.....)))))).))).....))))).))))...-..-...(((((....((((((......))).)))...))..))).... ( -24.10, z-score = -1.46, R) >droEre2.scaffold_4929 6285830 110 - 26641161 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC-ACAAAAGCCCAGGACGGUGUACGGAGCUAUGGCCAGAUGAAUGCAAA- .(((((...(((((......(((.((((((.....)))))).))).....)))))........-......(((..((.((.....))...))..)))))))).........- ( -23.10, z-score = 0.12, R) >droYak2.chr2L 14639761 110 - 22324452 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC-ACAAAAGCCCAGGACGGUAUACCAAGAUAUAGCCAGAUGAAUGCAAA- ..((((...(((((......(((.((((((.....)))))).))).....))))).))))...-...........((...((((......))))..)).............- ( -20.10, z-score = -0.83, R) >droSec1.super_3 793174 110 - 7220098 AUUUUGCCGGCUAAACCACACGAGCAGGUCAAAAUGAUAUGACCGCUAGGUGAGCGCAAACAC-ACAAAAGCCCAGGACGGUAUACGGAGAUACAGCCAGAUGAAUGCAAA- ..(((((.((((....(((...(((.(((((........))))))))..))).(.(......)-.)...))))..((...((((......))))..))........)))))- ( -25.60, z-score = -0.83, R) >droSim1.chr2L 14278135 110 - 22036055 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC-ACAAAAGCCCAGGACGGUAUACGGAGAUACAGCCAGAUGAAUGCAAA- ..((((...(((((......(((.((((((.....)))))).))).....))))).))))...-...........((...((((......))))..)).............- ( -21.60, z-score = -0.88, R) >droVir3.scaffold_12723 3152288 93 + 5802038 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCUCAAGCACUGCAUAGAUACAAAAUCAAGUACGAGUAUG------------------- .(((((((((((((......(((.((((((.....)))))).))).....)))))).............(((.....)))..)).)))))...------------------- ( -18.60, z-score = -0.65, R) >droMoj3.scaffold_6500 16482607 93 + 32352404 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAGCCGAGCUCAAGCACCGCACCGAUACAAAGCCAGGCACGAAUGCG------------------- ..(((((((((((........)))).)))))))......((((((...((.(.((....)).).))..)))).)).......(((....))).------------------- ( -19.70, z-score = -0.65, R) >droPer1.super_5 1897309 91 - 6813705 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCGCAAAC-----AGACACAAAACAGCGAGAGAGGAGAGAG---------------- ..((((...(((((......(((.((((((.....)))))).))).....)))))...)))).-----..........(..(....)..)......---------------- ( -18.30, z-score = -1.37, R) >dp4.chr4_group1 1954068 91 + 5278887 AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCGCAAAC-----AGACACAAAACAGCGAGAGAGGAGAGAG---------------- ..((((...(((((......(((.((((((.....)))))).))).....)))))...)))).-----..........(..(....)..)......---------------- ( -18.30, z-score = -1.37, R) >consensus AAUUUGACAGCUUAAACACACGAGCAUGUCAAAAUGAUAUGAUCGCUAAGUGAGCGCAAACAC_ACAAAAGCACAGAACGGUGUACGAAGAUACAGCCAGAUGAAUGCAAA_ ..((((...(((((......(((.((((((.....)))))).))).....))))).)))).................................................... (-14.34 = -14.62 + 0.28)

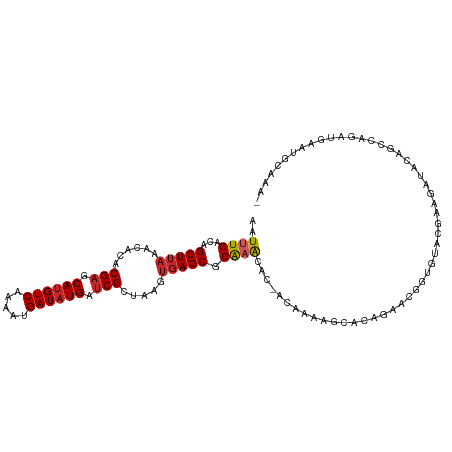
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:38 2011