
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,518,608 – 14,518,696 |
| Length | 88 |
| Max. P | 0.991924 |

| Location | 14,518,608 – 14,518,696 |
|---|---|
| Length | 88 |
| Sequences | 15 |
| Columns | 94 |
| Reading direction | forward |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.56481 |
| G+C content | 0.57515 |
| Mean single sequence MFE | -30.92 |
| Consensus MFE | -25.14 |
| Energy contribution | -24.47 |
| Covariance contribution | -0.67 |
| Combinations/Pair | 1.47 |
| Mean z-score | -1.37 |
| Structure conservation index | 0.81 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.29 |
| SVM RNA-class probability | 0.987620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
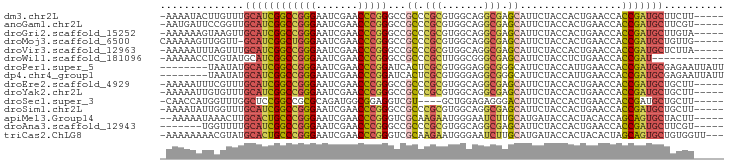
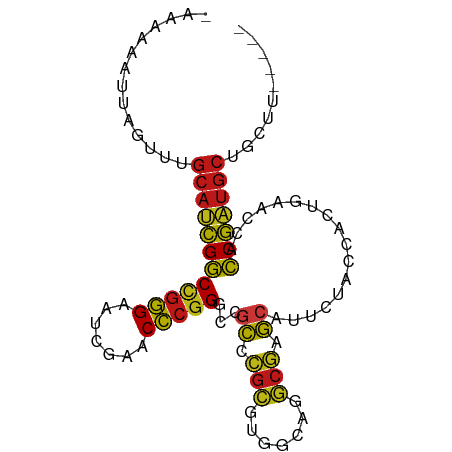
>dm3.chr2L 14518608 88 + 23011544 -AAAAUACUUGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCUU----- -.............((((((((((((.......)))))((((((.((...)).)))).)).................))))))).....----- ( -32.20, z-score = -1.90, R) >anoGam1.chr2L 14903949 88 + 48795086 -AAUGAUUCCGGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGU----- -........(((..((((((((((((.......)))))((((((.((...)).)))).)).................))))))).))).----- ( -33.80, z-score = -0.83, R) >droGri2.scaffold_15252 5781455 88 + 17193109 -AAAAAAGUAAGUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUGUA----- -.............((((((((((((.......)))))((((((.((...)).)))).)).................))))))).....----- ( -32.20, z-score = -1.42, R) >droMoj3.scaffold_6500 23190238 88 + 32352404 CAAAAAGUUGGUU-GCAUCGGCUGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUGUUG----- .............-((((((((((((.......)))))((((((.((...)).)))).)).................))))))).....----- ( -30.00, z-score = -0.27, R) >droVir3.scaffold_12963 204677 88 - 20206255 -AAAAAUUUAGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUCUUA----- -.............((((((((((((.......)))))((((((.((...)).)))).)).................))))))).....----- ( -32.20, z-score = -1.83, R) >droWil1.scaffold_181096 4521539 81 + 12416693 -AAAAACCUCGUAUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCUUGGCGGGCGAGCAUUCUACCUCUGAACCACCGAU------------ -...............((((((((((.......)))))(((((((((...))))))).)).................)))))------------ ( -31.40, z-score = -1.66, R) >droPer1.super_5 6190178 86 - 6813705 --------UAAUAUGCAUCGGCCGGGAAUCGAACCCGGAUCACUCGCGUGGGAGGCGGGCAUUCUACCAUUGAACCACCGAUGCGAGAAUUAUU --------.....(((((((((((((.......)))))....(((((.......)))))..................))))))))......... ( -27.80, z-score = -1.02, R) >dp4.chr4_group1 4588126 86 + 5278887 --------UAAUAUGCAUCGGCCGGGAAUCGAACCCGGAUCACUCGCGUGGGAGGCGGGCAUUCUACCAUUGAACCACCGAUGCGAGAAUUAUU --------.....(((((((((((((.......)))))....(((((.......)))))..................))))))))......... ( -27.80, z-score = -1.02, R) >droEre2.scaffold_4929 6276036 88 + 26641161 -AAAAAUUUCGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUGCUU----- -.........((..((((((((((((.......)))))((((((.((...)).)))).)).................))))))).))..----- ( -33.00, z-score = -1.46, R) >droYak2.chr2L 14628845 88 + 22324452 -AAAAAUUGUGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUGCUU----- -.........((..((((((((((((.......)))))((((((.((...)).)))).)).................))))))).))..----- ( -32.70, z-score = -1.22, R) >droSec1.super_3 781792 84 + 7220098 -CAACCAUGGUUUGGCUCCGGCCGCGCAGAUGGCGGAGGUCGU----GCUGGAGAGGGACAUUCUACCACUGAACCACCGAUGCUGCUU----- -.(((....))).(((....)))..((((....(((.(((((.----(.(((((((.....)))).)))))).))).)))...))))..----- ( -25.40, z-score = 0.82, R) >droSim1.chr2L 14267067 88 + 22036055 -AAAAUAUUGGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUGCUU----- -........(((..((((((((((((.......)))))((((((.((...)).)))).)).................))))))).))).----- ( -33.40, z-score = -1.35, R) >apiMel3.Group14 6137496 87 - 8318479 --AAAAAUAAACUUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUACCACUACACCAGCAGUGCUACUU----- --............((((((((((((.......)))))...((((((.......)))))).................))))))).....----- ( -29.30, z-score = -3.75, R) >droAna3.scaffold_12943 3039736 82 + 5039921 -------UGGUUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUUCGU----- -------.......((((((((((((.......)))))((((((.((...)).)))).)).................))))))).....----- ( -32.20, z-score = -0.89, R) >triCas2.ChLG8 3490820 90 - 15773733 -AAAAAAAACGUAUGCACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAUACCACUACACUAGCAGUGCUGUGGUU--- -........(((.......(((((((.......))))))).((((((.......)))))))))..(((((..(((.....)))..))))).--- ( -30.40, z-score = -2.74, R) >consensus _AAAAAAUUAGUUUGCAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCUACCACUGAACCACCGAUGCUGCUU_____ ..............((((((((((((.......)))))...((.(((.......))).)).................))))))).......... (-25.14 = -24.47 + -0.67)
| Location | 14,518,608 – 14,518,696 |
|---|---|
| Length | 88 |
| Sequences | 15 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 75.49 |
| Shannon entropy | 0.56481 |
| G+C content | 0.57515 |
| Mean single sequence MFE | -33.39 |
| Consensus MFE | -29.95 |
| Energy contribution | -28.58 |
| Covariance contribution | -1.37 |
| Combinations/Pair | 1.59 |
| Mean z-score | -1.59 |
| Structure conservation index | 0.90 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.51 |
| SVM RNA-class probability | 0.991924 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
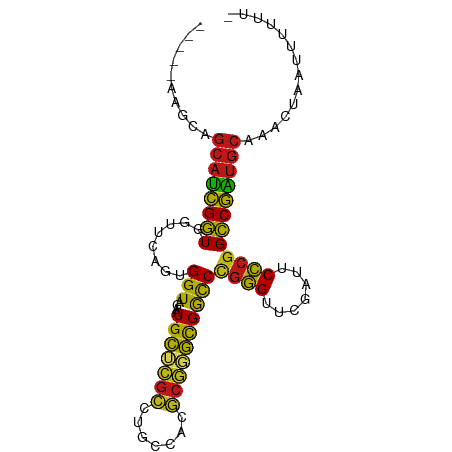
>dm3.chr2L 14518608 88 - 23011544 -----AAGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACAAGUAUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -1.84, R) >anoGam1.chr2L 14903949 88 - 48795086 -----ACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACCGGAAUCAUU- -----.(((....)))(((((((.(((((((.........)))))))(((..(((((.(((.......)))))))).))).....))))))).- ( -37.10, z-score = -1.04, R) >droGri2.scaffold_15252 5781455 88 - 17193109 -----UACAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAACUUACUUUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -2.01, R) >droMoj3.scaffold_6500 23190238 88 - 32352404 -----CAACAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCAGCCGAUGC-AACCAACUUUUUG -----.....((((((((.((((.......))))((.(((((((...)))))))))..(((.......))).))))))))-............. ( -33.70, z-score = -1.46, R) >droVir3.scaffold_12963 204677 88 + 20206255 -----UAAGAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACUAAAUUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -1.98, R) >droWil1.scaffold_181096 4521539 81 - 12416693 ------------AUCGGUGGUUCAGAGGUAGAAUGCUCGCCCGCCAAGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAUACGAGGUUUUU- ------------.(((...((((.......))))((((((((((...))))))((((.(((.......))))))))).))...))).......- ( -31.60, z-score = -0.43, R) >droPer1.super_5 6190178 86 + 6813705 AAUAAUUCUCGCAUCGGUGGUUCAAUGGUAGAAUGCCCGCCUCCCACGCGAGUGAUCCGGGUUCGAUUCCCGGCCGAUGCAUAUUA-------- ..........(((((((((((.....(((.....))).)))...(((....)))...((((.......))))))))))))......-------- ( -28.80, z-score = -1.24, R) >dp4.chr4_group1 4588126 86 - 5278887 AAUAAUUCUCGCAUCGGUGGUUCAAUGGUAGAAUGCCCGCCUCCCACGCGAGUGAUCCGGGUUCGAUUCCCGGCCGAUGCAUAUUA-------- ..........(((((((((((.....(((.....))).)))...(((....)))...((((.......))))))))))))......-------- ( -28.80, z-score = -1.24, R) >droEre2.scaffold_4929 6276036 88 - 26641161 -----AAGCAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACGAAAUUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -1.34, R) >droYak2.chr2L 14628845 88 - 22324452 -----AAGCAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACACAAUUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -1.58, R) >droSec1.super_3 781792 84 - 7220098 -----AAGCAGCAUCGGUGGUUCAGUGGUAGAAUGUCCCUCUCCAGC----ACGACCUCCGCCAUCUGCGCGGCCGGAGCCAAACCAUGGUUG- -----........(((((.((.((((((..((..(((..........----..))).))..))).))).)).)))))(((((.....))))).- ( -24.60, z-score = 0.44, R) >droSim1.chr2L 14267067 88 - 22036055 -----AAGCAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACCAAUAUUUU- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).............- ( -35.90, z-score = -1.50, R) >apiMel3.Group14 6137496 87 + 8318479 -----AAGUAGCACUGCUGGUGUAGUGGUAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAAGUUUAUUUUU-- -----.....((((((((((((......)))))(((((((.......)))))))..(((((.......))))))))))))............-- ( -32.20, z-score = -3.39, R) >droAna3.scaffold_12943 3039736 82 - 5039921 -----ACGAAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAAACCA------- -----.....(((((((..((((.......))))((.(((((((...)))))))))(((((.......)))))))))))).......------- ( -35.90, z-score = -1.36, R) >triCas2.ChLG8 3490820 90 + 15773733 ---AACCACAGCACUGCUAGUGUAGUGGUAUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUGCAUACGUUUUUUUU- ---.(((((.((((.....)))).))))).....((((((.......))))))(.((((((.......)))))))..................- ( -32.70, z-score = -3.86, R) >consensus _____AAGCAGCAUCGGUGGUUCAGUGGUAGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUGCAAACUAAUUUUUU_ ..........((((((((........(((....(((((((.......))))))))))((((.......)))))))))))).............. (-29.95 = -28.58 + -1.37)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:36 2011