| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,518,087 – 14,518,159 |
| Length | 72 |
| Max. P | 0.901079 |

| Location | 14,518,087 – 14,518,159 |
|---|---|
| Length | 72 |
| Sequences | 13 |
| Columns | 78 |
| Reading direction | forward |
| Mean pairwise identity | 84.19 |
| Shannon entropy | 0.34414 |
| G+C content | 0.63926 |
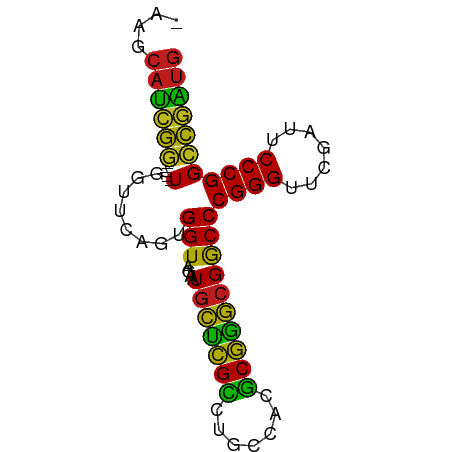
| Mean single sequence MFE | -29.85 |
| Consensus MFE | -26.18 |
| Energy contribution | -24.26 |
| Covariance contribution | -1.92 |
| Combinations/Pair | 1.67 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.88 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.99 |
| SVM RNA-class probability | 0.868769 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
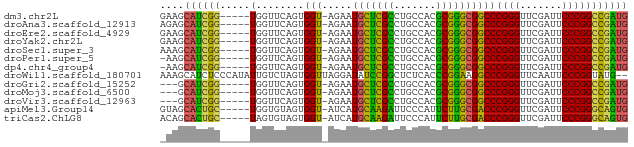
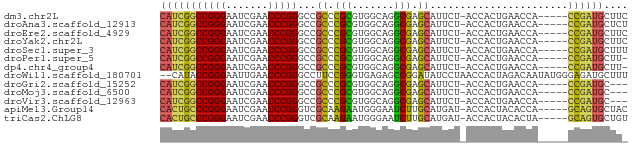
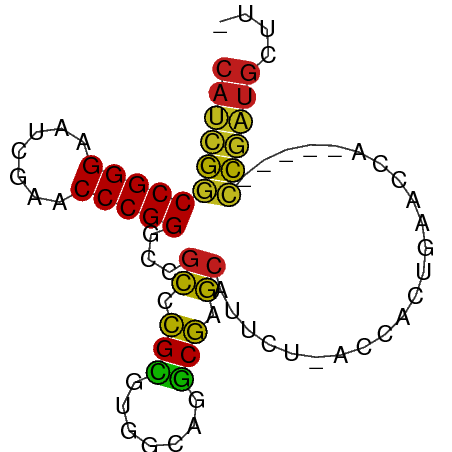
>dm3.chr2L 14518087 72 + 23011544 GAAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.82, R) >droAna3.scaffold_12913 128343 72 - 441482 AGAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.73, R) >droEre2.scaffold_4929 6275463 72 + 26641161 GAAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.82, R) >droYak2.chr2L 14627550 72 + 22324452 GAAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.82, R) >droSec1.super_3 781237 72 + 7220098 AAAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.94, R) >droPer1.super_5 6189145 71 - 6813705 -AAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG -...((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.94, R) >dp4.chr4_group4 1436228 71 + 6586962 -AAGCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG -...((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.94, R) >droWil1.scaffold_180701 2121832 76 - 3904529 AAAGCAUCUCCCAUAUUGUCUAGUGGUUAGGAUAUCCGGCUCUCACCCGGAAGGCCCGGGUUCAAUUCCCGGUAUG-- ...((((((.((((........))))...)))).(((((.......)))))..))(((((.......)))))....-- ( -21.50, z-score = -0.44, R) >droGri2.scaffold_15252 11469071 69 + 17193109 ---GCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ---.((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.99, R) >droMoj3.scaffold_6500 23189631 69 + 32352404 ---GCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ---.((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.99, R) >droVir3.scaffold_12963 203613 69 - 20206255 ---GCAUCGG-----UGGUUCAGUGGU-AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ---.((((((-----..((((......-.))))((.(((((((...)))))))))(((((.......))))))))))) ( -30.80, z-score = -0.99, R) >apiMel3.Group14 2180898 72 + 8318479 GUAGCACUGC-----UGGUGUAGUGGU-AUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUG (((.((((((-----....)))))).)-))...((((((.......))))))(.((((((.......))))))).... ( -30.20, z-score = -3.23, R) >triCas2.ChLG8 12282827 72 + 15773733 ACAGCACUGC-----UAGUGUAGUGGU-AUCAUGCAAGAUUCCCAUUCUUGCGACCCGGGUUCGAUUCCCGGGCAGUG ....((((((-----....))))))..-.....((((((.......))))))(.((((((.......))))))).... ( -28.40, z-score = -3.19, R) >consensus _AAGCAUCGG_____UGGUUCAGUGGU_AGAAUGCUCGCCUGCCACGCGGGCGGCCCGGGUUCGAUUCCCGGCCGAUG ....((((((.......((((........))))((((((.......))))))...(((((.......))))))))))) (-26.18 = -24.26 + -1.92)
| Location | 14,518,087 – 14,518,159 |
|---|---|
| Length | 72 |
| Sequences | 13 |
| Columns | 78 |
| Reading direction | reverse |
| Mean pairwise identity | 84.19 |
| Shannon entropy | 0.34414 |
| G+C content | 0.63926 |
| Mean single sequence MFE | -27.18 |
| Consensus MFE | -22.39 |
| Energy contribution | -21.22 |
| Covariance contribution | -1.17 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.51 |
| Structure conservation index | 0.82 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.15 |
| SVM RNA-class probability | 0.901079 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14518087 72 - 23011544 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUUC (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).... ( -28.00, z-score = -1.31, R) >droAna3.scaffold_12913 128343 72 + 441482 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUCU (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).... ( -28.00, z-score = -1.21, R) >droEre2.scaffold_4929 6275463 72 - 26641161 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUUC (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).... ( -28.00, z-score = -1.31, R) >droYak2.chr2L 14627550 72 - 22324452 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUUC (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).... ( -28.00, z-score = -1.31, R) >droSec1.super_3 781237 72 - 7220098 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUUU (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).... ( -28.00, z-score = -1.33, R) >droPer1.super_5 6189145 71 + 6813705 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUU- (((((((((((.......)))))((((((.((...)).)))).)).....-............-----))))))...- ( -28.00, z-score = -1.36, R) >dp4.chr4_group4 1436228 71 - 6586962 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGCUU- (((((((((((.......)))))((((((.((...)).)))).)).....-............-----))))))...- ( -28.00, z-score = -1.36, R) >droWil1.scaffold_180701 2121832 76 + 3904529 --CAUACCGGGAAUUGAACCCGGGCCUUCCGGGUGAGAGCCGGAUAUCCUAACCACUAGACAAUAUGGGAGAUGCUUU --....(((((.......)))))(((((((((((....)))((....))................))))))..))... ( -23.20, z-score = -0.99, R) >droGri2.scaffold_15252 11469071 69 - 17193109 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGC--- (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).--- ( -28.00, z-score = -1.47, R) >droMoj3.scaffold_6500 23189631 69 - 32352404 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGC--- (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).--- ( -28.00, z-score = -1.47, R) >droVir3.scaffold_12963 203613 69 + 20206255 CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU-ACCACUGAACCA-----CCGAUGC--- (((((((((((.......)))))((((((.((...)).)))).)).....-............-----)))))).--- ( -28.00, z-score = -1.47, R) >apiMel3.Group14 2180898 72 - 8318479 CACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAU-ACCACUACACCA-----GCAGUGCUAC (((((((((((.......)))))...((((((.......)))))).....-............-----)))))).... ( -25.10, z-score = -2.70, R) >triCas2.ChLG8 12282827 72 - 15773733 CACUGCCCGGGAAUCGAACCCGGGUCGCAAGAAUGGGAAUCUUGCAUGAU-ACCACUACACUA-----GCAGUGCUGU (((((((((((.......)))))...((((((.......)))))).....-............-----)))))).... ( -25.10, z-score = -2.32, R) >consensus CAUCGGCCGGGAAUCGAACCCGGGCCGCCCGCGUGGCAGGCGAGCAUUCU_ACCACUGAACCA_____CCGAUGCUU_ (((((((((((.......)))))...((.(((.......))).)).......................)))))).... (-22.39 = -21.22 + -1.17)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:34 2011