
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,507,132 – 14,507,237 |
| Length | 105 |
| Max. P | 0.811691 |

| Location | 14,507,132 – 14,507,234 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.83 |
| Shannon entropy | 0.28264 |
| G+C content | 0.33366 |
| Mean single sequence MFE | -23.67 |
| Consensus MFE | -16.21 |
| Energy contribution | -15.81 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.91 |
| Structure conservation index | 0.68 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.52 |
| SVM RNA-class probability | 0.726477 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
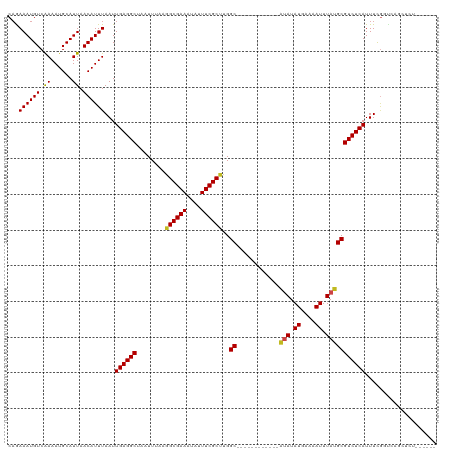
>dm3.chr2L 14507132 102 + 23011544 CAAAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU------ ...((((((.((......)).))))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..............------ ( -23.10, z-score = -2.24, R) >droSim1.chr2L 14254691 102 + 22036055 CAAAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU------ ...((((((.((......)).))))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..............------ ( -23.10, z-score = -2.24, R) >droSec1.super_3 770367 102 + 7220098 CAAAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU------ ...((((((.((......)).))))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..............------ ( -23.10, z-score = -2.24, R) >droYak2.chr2L 14615982 102 + 22324452 CAAAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU------ ...((((((.((......)).))))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..............------ ( -25.80, z-score = -3.05, R) >droEre2.scaffold_4929 6261404 101 + 26641161 CAAAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGC-UGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU------ ...((((((.((......)).))))))...((((((........((((((....)))))-).((------------(((.((....)).)))))))))))..............------ ( -22.50, z-score = -2.08, R) >droAna3.scaffold_12916 14869352 102 - 16180835 CAGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGCCAGAUCC------ ..(((((....(((((......)))))....(((((.......(((((((....))))))).((------------(((.((....)).)))))))))))))))..........------ ( -24.90, z-score = -2.20, R) >dp4.chr4_group1 1923611 120 - 5278887 CAGAAAUGUCGAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCCGGCUCAUACAUGCGAGUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAAAAAAUGCUCA ((....)).......(((((...((..((.((((((........((((((....))))))..((........))..(((((........))))))))))).))..)).....)))))... ( -24.60, z-score = -0.36, R) >droPer1.super_5 1868122 120 + 6813705 CAGAAAUGUCGAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCCGGCUCAUACAUGCGAGUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAAAAAAUGCUCA ((....)).......(((((...((..((.((((((........((((((....))))))..((........))..(((((........))))))))))).))..)).....)))))... ( -24.60, z-score = -0.36, R) >droWil1.scaffold_180764 2825537 102 + 3949147 CAGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUUAAGUCUUUAC------ ....(((((......)))))...((((((.((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..))))))......------ ( -25.00, z-score = -3.07, R) >droVir3.scaffold_12723 2672223 92 + 5802038 CAGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------AUAUAUGAGAAUUUAUGCGUCUCAUUUU---------------- ...((((((.((......)).))))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))....---------------- ( -24.40, z-score = -3.09, R) >droMoj3.scaffold_6500 15901159 98 + 32352404 CCGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------UCAUAUGAGAAUUUAUGCGUCUCAUUUUGUCGCA---------- .(((((((.....((((((.....((((..((((.(.......(((((((....)))))))).)------------)))..)))).....))))))..))))))).....---------- ( -24.11, z-score = -1.80, R) >droGri2.scaffold_15126 7553577 98 - 8399593 CAGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUCGGC------------ACAUAUGAGAAUUUAUGCGUCUCAUUUUGUCGCA---------- ...((((((.((......)).)))))).......((........((((((....)))))).(((------------(...((((((.........))))))..)))))).---------- ( -18.80, z-score = -0.17, R) >consensus CAGAAAUGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC____________AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGCUCU______ ...((((((.((......)).))))))...((((((........((((((....)))))).................((((........)))).)))))).................... (-16.21 = -15.81 + -0.40)


| Location | 14,507,138 – 14,507,237 |
|---|---|
| Length | 99 |
| Sequences | 12 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.01 |
| Shannon entropy | 0.32151 |
| G+C content | 0.34565 |
| Mean single sequence MFE | -23.27 |
| Consensus MFE | -15.36 |
| Energy contribution | -15.10 |
| Covariance contribution | -0.26 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.93 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811691 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
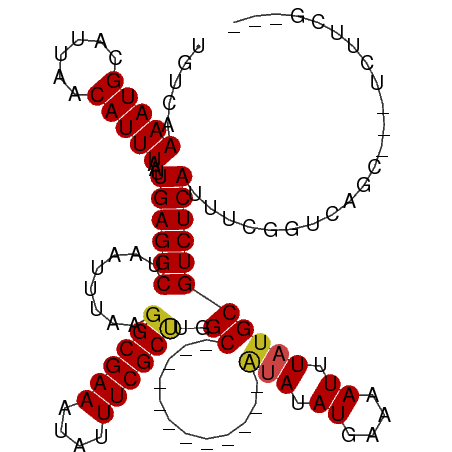
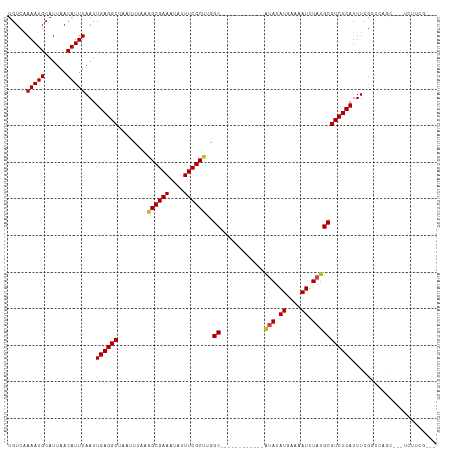
>dm3.chr2L 14507138 99 + 23011544 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC---UCUUCG--- ((.(.(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).))))))))))).....).))..---......--- ( -22.70, z-score = -2.14, R) >droSim1.chr2L 14254697 99 + 22036055 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC---UCUUCG--- ((.(.(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).))))))))))).....).))..---......--- ( -22.70, z-score = -2.14, R) >droSec1.super_3 770373 99 + 7220098 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC---UCUUCG--- ((.(.(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).))))))))))).....).))..---......--- ( -22.70, z-score = -2.14, R) >droYak2.chr2L 14615988 99 + 22324452 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC---UCUUCG--- ((.(.(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).))))))))))).....).))..---......--- ( -25.40, z-score = -2.94, R) >droEre2.scaffold_4929 6261410 98 + 26641161 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGC-UGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC---UCUUCG--- ((.(.(((((......)))))...((((((........((((((....)))))-).((------------(((.((....)).))))))))))).....).))..---......--- ( -22.10, z-score = -1.93, R) >droAna3.scaffold_12916 14869358 99 - 16180835 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGCCAGA---UCCUCG--- .(((.(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))....)))....---......--- ( -24.70, z-score = -2.38, R) >dp4.chr4_group1 1923617 117 - 5278887 UGUCGAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCCGGCUCAUACAUGCGAGUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAAAAAAUGCUCACGG ...((....(((((...((..((.((((((........((((((....))))))..((........))..(((((........))))))))))).))..)).....)))))...)). ( -25.00, z-score = -0.61, R) >droPer1.super_5 1868128 117 + 6813705 UGUCGAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCCGGCUCAUACAUGCGAGUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAAAAAAUGCUCACGG ...((....(((((...((..((.((((((........((((((....))))))..((........))..(((((........))))))))))).))..)).....)))))...)). ( -25.00, z-score = -0.61, R) >droWil1.scaffold_180764 2825543 99 + 3949147 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC------------AUAUAUGAAAAUUUAUGCGUCUCAUUUUAAGUCUUUACGGU------ .................((((((.((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))..)))))).........------ ( -23.30, z-score = -2.61, R) >droVir3.scaffold_12723 2672229 86 + 5802038 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------AUAUAUGAGAAUUUAUGCGUCUCAUUUU------------------- .....(((((......)))))...((((((.......(((((((....))))))).((------------(((.((....)).)))))))))))....------------------- ( -23.70, z-score = -3.26, R) >droMoj3.scaffold_6500 15901165 95 + 32352404 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCCUGGC------------UCAUAUGAGAAUUUAUGCGUCUCAUUUUGUCGCAUCU---------- .......((((....(((......((((((.......(((((((....)))))))..(------------((....))).........))))))...))).))))..---------- ( -22.10, z-score = -1.56, R) >droGri2.scaffold_15126 7553583 98 - 8399593 UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUCGGC------------ACAUAUGAGAAUUUAUGCGUCUCAUUUUGUCGCAUCUCUU------- .....(((((......)))))....((((.........((((((....))))))((((------------(...((((((.........))))))..)))))..))))..------- ( -19.90, z-score = -0.86, R) >consensus UGUCAAAAUGCAUUAACAUUUAAUUGAGGCUAAUUUAAGGCGAAAUAUUUCGCUUGGC____________AUAUAUGAAAAUUUAUGCGUCUCAUUUCGGUCAGC___UCUUCG___ .....(((((......)))))...((((((........((((((....)))))).................((((........)))).))))))....................... (-15.36 = -15.10 + -0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:31 2011