
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,505,467 – 14,505,558 |
| Length | 91 |
| Max. P | 0.752822 |

| Location | 14,505,467 – 14,505,558 |
|---|---|
| Length | 91 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 74.60 |
| Shannon entropy | 0.49239 |
| G+C content | 0.42039 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -10.06 |
| Energy contribution | -10.46 |
| Covariance contribution | 0.40 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.44 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.752822 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
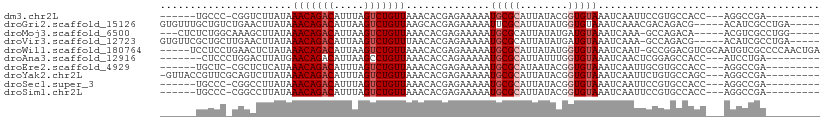
>dm3.chr2L 14505467 91 - 23011544 ------UGCCC-CGGUCUUAUAAACAGACAUUUAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUACGGUGUAAAUCAAUUCCGUGCCACC---AGGCCGA--------- ------.....-((((((....(((((((.....)))))))...((((.(((...(((((........)))))......))))))).....---)))))).--------- ( -25.10, z-score = -2.74, R) >droGri2.scaffold_15126 843122 100 - 8399593 GUGUUUGCUGUCUGAACUUAUAAACAGACAUUAAGUCUGUUAAGCACGAGAAAAAUUCGCAUUAUAUGGUGUAAAUCAAACGACAGACG-----ACAUCGCCUGA----- (((..((.((((((........(((((((.....)))))))..((((.(.................).))))...........))))))-----.)).)))....----- ( -21.53, z-score = -0.77, R) >droMoj3.scaffold_6500 16447569 96 + 32352404 ---CUCUCUGGCAAAGCUUAUAAACAGACAUUAAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUAUGAUGUAAAUCAAA-GCCAGACA-----ACGUCGCCUGG----- ---...((((((..........(((((((.....))))))).................(((((....)))))........-))))))..-----...........----- ( -21.40, z-score = -2.00, R) >droVir3.scaffold_12723 3127500 99 + 5802038 GUGUUCGCUGCUUGAACUUAUAAACAGACAUUAAGUCUGUUUAACACGAGAAAAAUGCGCAUUAUAUGAUGUAAAUCAAA-GCCAGACG-----ACAUCGCCUGA----- (((.(((((((((((.....(((((((((.....)))))))))...............(((((....)))))...)).))-).))).))-----)...)))....----- ( -22.90, z-score = -1.79, R) >droWil1.scaffold_180764 1126924 104 + 3949147 -----UCCUCCUGAACUCUAUAAACAGACAUUAAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUAUGGUGUAAAUCAAU-GCCGGACGUCGCAAUGUCGCCCCAACUGA -----..........(((....(((((((.....)))))))......)))........((......((((((......))-))))(((((....)))))))......... ( -19.30, z-score = -0.56, R) >droAna3.scaffold_12916 14867875 91 + 16180835 -------CUCCCUGGACUUAUGAACAGACAUUAAGCCUGUUAAACACCAGAAAAAUGCGCAUUAUUUGGUGUAAAUCAACUCGGAGCCACC---AUCCUGA--------- -------((((((((.......(((((.(.....).))))).....)))).....(((((........))))).........)))).....---.......--------- ( -16.90, z-score = -0.58, R) >droEre2.scaffold_4929 6259814 91 - 26641161 ------UGCUC-CGCUCUCAUAAACAGACAUUUAGUCUGUUAAACACGAGAAAAAUGCGCAUAAUACGGUGUAAAUCAAUUGCGUGCCACC---AGGCCGA--------- ------.....-(((((((...(((((((.....)))))))......))))....(((((........)))))........))).(((...---.)))...--------- ( -21.00, z-score = -1.08, R) >droYak2.chr2L 14614299 97 - 22324452 -GUUACCGUUCGCAGUCUUAUAAACAGACAUUUAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUACGGUGUAAAUCAAUUCUGUGCCAGC---AGGCCGA--------- -..((((((.((((.((((...(((((((.....)))))))......))))....))))......))))))...........((.(((...---.))))).--------- ( -24.30, z-score = -1.80, R) >droSec1.super_3 768716 91 - 7220098 ------UGCCC-CGGCCUUAUAAACAGACAUUUAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUACGGUGUAAAUCAAUUCCGUGCCACC---AGGCCGA--------- ------.....-((((((....(((((((.....)))))))...((((.(((...(((((........)))))......))))))).....---)))))).--------- ( -27.80, z-score = -3.65, R) >droSim1.chr2L 14253041 91 - 22036055 ------UGCCC-CGGCCUUAUAAACAGACAUUUAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUACGGUGUAAAUCAAUUCCGUGCCACC---AGGCCGA--------- ------.....-((((((....(((((((.....)))))))...((((.(((...(((((........)))))......))))))).....---)))))).--------- ( -27.80, z-score = -3.65, R) >consensus ______UCUCC_CGGACUUAUAAACAGACAUUAAGUCUGUUAAACACGAGAAAAAUGCGCAUUAUACGGUGUAAAUCAAUUCCGUGCCACC___AGGCCGA_________ ......................(((((((.....)))))))..............(((((........)))))..................................... (-10.06 = -10.46 + 0.40)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:29 2011