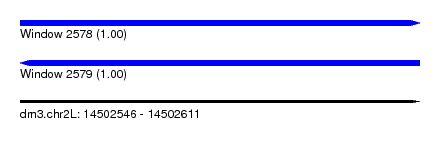
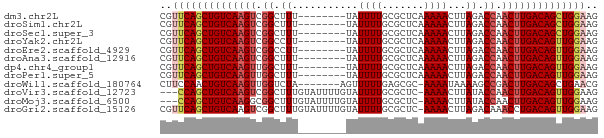
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,502,546 – 14,502,611 |
| Length | 65 |
| Max. P | 0.999674 |

| Location | 14,502,546 – 14,502,611 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 73 |
| Reading direction | forward |
| Mean pairwise identity | 85.56 |
| Shannon entropy | 0.29527 |
| G+C content | 0.43049 |
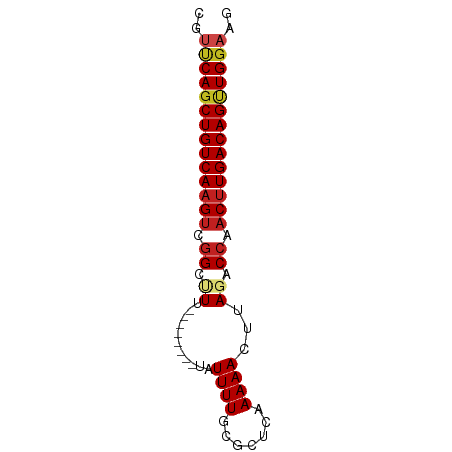
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -18.50 |
| Energy contribution | -18.63 |
| Covariance contribution | 0.13 |
| Combinations/Pair | 1.13 |
| Mean z-score | -5.09 |
| Structure conservation index | 0.72 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.17 |
| SVM RNA-class probability | 0.999674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
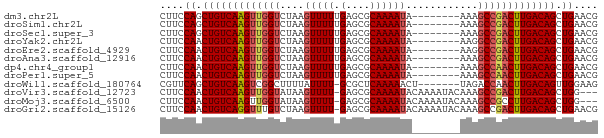
>dm3.chr2L 14502546 65 + 23011544 CUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCGACUUGACAGCUGAACG ....((((((((((((((((......(((((....)))))..--------...)))))))))))))))).... ( -30.30, z-score = -6.50, R) >droSim1.chr2L 14249789 65 + 22036055 CUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCGACUUGACAGCUGAACG ....((((((((((((((((......(((((....)))))..--------...)))))))))))))))).... ( -30.30, z-score = -6.50, R) >droSec1.super_3 765771 65 + 7220098 CUUCCAGCUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCGACUUGACAGCUGAACG ....((((((((((((((((......(((((....)))))..--------...)))))))))))))))).... ( -30.30, z-score = -6.50, R) >droYak2.chr2L 14611312 65 + 22324452 CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAGGCCGACUUGACAGCUGAACG ....((.(((((((((((((((....(((((....)))))..--------.))))))))))))))).)).... ( -27.70, z-score = -5.65, R) >droEre2.scaffold_4929 6257049 65 + 26641161 CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAGGCCGACUUGACAGCUGAACG ....((.(((((((((((((((....(((((....)))))..--------.))))))))))))))).)).... ( -27.70, z-score = -5.65, R) >droAna3.scaffold_12916 14864911 65 - 16180835 CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCGACUUGACAGCUGAACG ....((.(((((((((((((......(((((....)))))..--------...))))))))))))).)).... ( -23.70, z-score = -4.58, R) >dp4.chr4_group1 1910345 65 - 5278887 CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCAACUUGACAGCUGAACG ....((.(((((((((((((......(((((....)))))..--------...))))))))))))).)).... ( -24.00, z-score = -4.99, R) >droPer1.super_5 1849355 65 + 6813705 CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA--------AAAGCCAACUUGACAGCUGAACG ....((.(((((((((((((......(((((....)))))..--------...))))))))))))).)).... ( -24.00, z-score = -4.99, R) >droWil1.scaffold_180764 2818868 65 + 3949147 CGUUCAGCUGUCAAGUCGGCUUUUAUUUU-GCGCUCAAAAACU-------UAGACCAACUUGACAGUUGGAAG ..((((((((((((((.((((....((((-(....)))))...-------.)).)).)))))))))))))).. ( -25.20, z-score = -4.89, R) >droVir3.scaffold_12723 3124526 69 - 5802038 CUUCCAACUGUCAAGUUGGUAUAAGUUUU-GAGCGCAAAAUACAAAAUACAAAGCCGACUUGACAGCUGG--- ...(((.(((((((((((((....(((((-(...........)))))).....))))))))))))).)))--- ( -25.40, z-score = -5.44, R) >droMoj3.scaffold_6500 16442255 69 - 32352404 CUUCCAACUGUCAAGUUGGUAUAAGUUUU-GAGCGCAAAAUACAAAAUACAAAGCCGCCUUGACAGCUGG--- ...(((.((((((((.((((....(((((-(...........)))))).....)))).)))))))).)))--- ( -20.80, z-score = -3.36, R) >droGri2.scaffold_15126 839420 72 + 8399593 CUUCCAACUGUCAGGUUUGUCUAAGUUUU-GAGCGCAAAAUACAAAAUACAAAGCCGACUUGACAGCUGAACG ....((.((((((((((.((....(((((-(...........)))))).....)).)))))))))).)).... ( -17.30, z-score = -1.99, R) >consensus CUUCCAACUGUCAAGUUGGUCUAAGUUUUUGAGCGCAAAAUA________AAAGCCGACUUGACAGCUGAACG ....((.(((((((((((((.................................))))))))))))).)).... (-18.50 = -18.63 + 0.13)
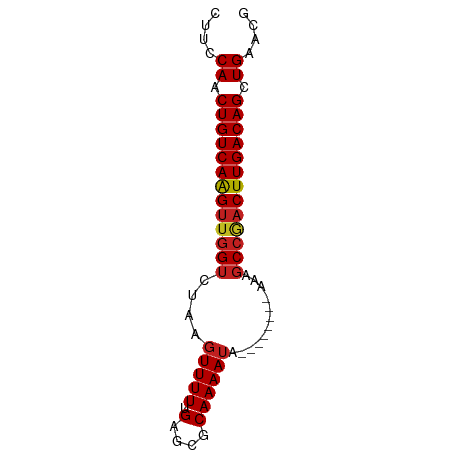
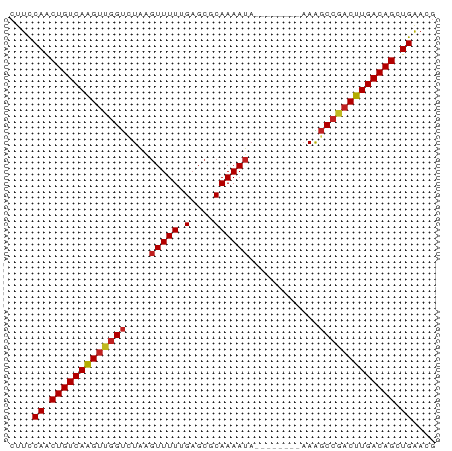
| Location | 14,502,546 – 14,502,611 |
|---|---|
| Length | 65 |
| Sequences | 12 |
| Columns | 73 |
| Reading direction | reverse |
| Mean pairwise identity | 85.56 |
| Shannon entropy | 0.29527 |
| G+C content | 0.43049 |
| Mean single sequence MFE | -25.67 |
| Consensus MFE | -19.09 |
| Energy contribution | -19.38 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.12 |
| Mean z-score | -4.92 |
| Structure conservation index | 0.74 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 4.11 |
| SVM RNA-class probability | 0.999637 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14502546 65 - 23011544 CGUUCAGCUGUCAAGUCGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAG ..((((((((((((((.((((..--------..(((((....)))))....)).)).)))))))))))))).. ( -27.60, z-score = -5.56, R) >droSim1.chr2L 14249789 65 - 22036055 CGUUCAGCUGUCAAGUCGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAG ..((((((((((((((.((((..--------..(((((....)))))....)).)).)))))))))))))).. ( -27.60, z-score = -5.56, R) >droSec1.super_3 765771 65 - 7220098 CGUUCAGCUGUCAAGUCGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGCUGGAAG ..((((((((((((((.((((..--------..(((((....)))))....)).)).)))))))))))))).. ( -27.60, z-score = -5.56, R) >droYak2.chr2L 14611312 65 - 22324452 CGUUCAGCUGUCAAGUCGGCCUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..((((((((((((((.((.((.--------..(((((....)))))....)).)).)))))))))))))).. ( -24.80, z-score = -4.72, R) >droEre2.scaffold_4929 6257049 65 - 26641161 CGUUCAGCUGUCAAGUCGGCCUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..((((((((((((((.((.((.--------..(((((....)))))....)).)).)))))))))))))).. ( -24.80, z-score = -4.72, R) >droAna3.scaffold_12916 14864911 65 + 16180835 CGUUCAGCUGUCAAGUCGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..((((((((((((((.((((..--------..(((((....)))))....)).)).)))))))))))))).. ( -25.20, z-score = -4.89, R) >dp4.chr4_group1 1910345 65 + 5278887 CGUUCAGCUGUCAAGUUGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..(((((((((((((((((((..--------..(((((....)))))....)).))))))))))))))))).. ( -29.30, z-score = -6.35, R) >droPer1.super_5 1849355 65 - 6813705 CGUUCAGCUGUCAAGUUGGCUUU--------UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..(((((((((((((((((((..--------..(((((....)))))....)).))))))))))))))))).. ( -29.30, z-score = -6.35, R) >droWil1.scaffold_180764 2818868 65 - 3949147 CUUCCAACUGUCAAGUUGGUCUA-------AGUUUUUGAGCGC-AAAAUAAAAGCCGACUUGACAGCUGAACG ....((.(((((((((((((...-------...(((((....)-)))).....))))))))))))).)).... ( -23.70, z-score = -4.58, R) >droVir3.scaffold_12723 3124526 69 + 5802038 ---CCAGCUGUCAAGUCGGCUUUGUAUUUUGUAUUUUGCGCUC-AAAACUUAUACCAACUUGACAGUUGGAAG ---(((((((((((((.((...((..(((((...........)-))))..))..)).)))))))))))))... ( -24.90, z-score = -4.94, R) >droMoj3.scaffold_6500 16442255 69 + 32352404 ---CCAGCUGUCAAGGCGGCUUUGUAUUUUGUAUUUUGCGCUC-AAAACUUAUACCAACUUGACAGUUGGAAG ---((((((((((((..((...((..(((((...........)-))))..))..))..))))))))))))... ( -23.70, z-score = -3.87, R) >droGri2.scaffold_15126 839420 72 - 8399593 CGUUCAGCUGUCAAGUCGGCUUUGUAUUUUGUAUUUUGCGCUC-AAAACUUAGACAAACCUGACAGUUGGAAG ..(((((((((((.((..(.((((..(((((...........)-))))..)))))..)).))))))))))).. ( -19.50, z-score = -1.95, R) >consensus CGUUCAGCUGUCAAGUCGGCUUU________UAUUUUGCGCUCAAAAACUUAGACCAACUUGACAGUUGGAAG ..((((((((((((((.((...................................)).)))))))))))))).. (-19.09 = -19.38 + 0.29)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:28 2011