| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,472,799 – 14,472,929 |
| Length | 130 |
| Max. P | 0.688134 |

| Location | 14,472,799 – 14,472,929 |
|---|---|
| Length | 130 |
| Sequences | 10 |
| Columns | 160 |
| Reading direction | forward |
| Mean pairwise identity | 67.52 |
| Shannon entropy | 0.59010 |
| G+C content | 0.54377 |
| Mean single sequence MFE | -37.07 |
| Consensus MFE | -17.16 |
| Energy contribution | -17.91 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.42 |
| SVM RNA-class probability | 0.688134 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
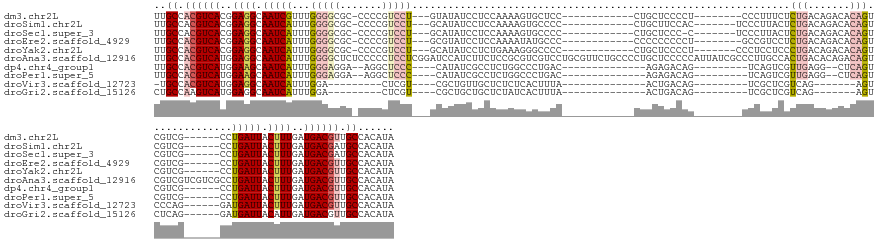
>dm3.chr2L 14472799 130 + 23011544 UUGCCACGUCACGGAGGCAAUCGUUUGGGGCGC-CCCCGUCCU---GUAUAUCCUCCAAAAGUGCUCC------------CUGCUCCCCU--------CCCUUUCUCUGACAGACACAGUCGUCG------CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.((((((.(((((..((.(..(((((...-)))))..).---))....))))).(((((((...------------..))......--------.....((..((((.(((...)))))))------...))..)))))..)))))).))...... ( -32.90, z-score = -0.56, R) >droSim1.chr2L 14218689 131 + 22036055 UUGCCACGUCACGGAGGCAAUCAUUUGGGGCGC-CCCCGUCCU---GCAUAUCCUCCAAAAGUGCCCC------------CUGCUUCCAC-------UCCCUUACUCUGACAGACACAGUCGUCG------CCUGAUUACUUUGAUGACGAUGCCACAUA .((.(((((((.(((((((.......(((((((-.........---...............)))))))------------.)))))))..-------.................((.(((.(((.------...))).))).)).))))).)).)).... ( -31.76, z-score = -0.47, R) >droSec1.super_3 728948 130 + 7220098 UUGCCACGUCACGGAGGCAAUCAUUUGGGGCGC-CCCCGUCCU---GCAUAUCCUCCAAAAGUGCCCC------------CUGCUCCC-C-------UCCCUUACUCUGACAGACACAGUCGUCG------CCUGAUUACUUUGAUGACGAUGCCACAUA .((.((((((((((((..(((((...(((((((-.........---...............)))))))------------........-.-------...........(((.(((...)))))).------..))))).))))).))))).)).)).... ( -31.26, z-score = -0.27, R) >droEre2.scaffold_4929 6225152 130 + 26641161 UUGCCACGUCACGGAGGCAAUCAUUUGGGGCGC-CCCCGUCCU---GCGUAUCCUCCAAAAUAUGCCC------------CCCCCCCCCU--------GCCGUCCUCUGACAGACACAGUCGUCG------CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.((((((.(((((((.......((((((.-...))))))---((((((........))))))..------------.........)--------))).))).........((.(((.(((.------...))).))).)).)))))).))...... ( -35.40, z-score = -1.35, R) >droYak2.chr2L 14580277 131 + 22324452 UUGCCACGUCACGGAGGCAAUCAUUUGGGGCGC-CCCCGUCCU---GCAUAUCCUCUGAAAGGGCCCC------------CUGCUCCCCU-------CCCUCCUCCCUGACAGACACAGUCGUCG------CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.((((((.(((((.........((((.((-....(((((---..............)))))...------------..)).)))).-------....))))).)))).......(((((((------...........))))))))).))...... ( -38.28, z-score = -1.17, R) >droAna3.scaffold_12916 14828343 160 - 16180835 UUGCCACGUCAUGGAGGCAAUCAUUUGGGGCUCUCCCCCUCCUCGGAUCCAUCUUCUCCGCGUCGUCCUGCGUUCUGCCCCUGCUCCCCCAUUAUCGCCCUUGCCACUGACACAGACAGUCGUCGUCGUCGCCUGAUUACUUUGAUGACGUUGCCACAUA ..((.(((((((.((((.(((((...(((((..(((........)))...........((((......))))....))))).((............((....))....(((((.(((....))))).))))).))))).)))).))))))).))...... ( -42.90, z-score = -0.79, R) >dp4.chr4_group1 1862945 123 - 5278887 UUGCCACGUCAUGGAAGCAAUCAUUUGGGAGGA--AGGCUCCC----CAUAUCGCCUCUGGCCCUGAC--------------AGAGACAG---------UCAGUCGUUGAGG--CUCAGUCGUCG------CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.(((((((.((((.(((((...(((((..--...)))))----......(((((.(((.(((((--------------.......)---------))))..)))))))--)..........------..))))).)))).))))))).))...... ( -44.70, z-score = -2.15, R) >droPer1.super_5 1803078 123 + 6813705 UUGCCACGUCAUGGAAGCAAUCAUUUGGGAGGA--AGGCUCCC----CAUAUCGCCUCUGGCCCUGAC--------------AGAGACAG---------UCAGUCGUUGAGG--CUCAGUCGUCG------CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.(((((((.((((.(((((...(((((..--...)))))----......(((((.(((.(((((--------------.......)---------))))..)))))))--)..........------..))))).)))).))))))).))...... ( -44.70, z-score = -2.15, R) >droVir3.scaffold_12723 3089450 110 - 5802038 -UGCCACGUCAUGGAGGCAAUCAUUUGGA---------CUCGU----CGCUGUUGCUCUCUCACUUUA--------------ACUGACAG---------UCGCUCGUCAG-------AGUCCCAG------GAUGAUUACUUUGAUGACGUUGCCACAUA -.((.(((((((.((((.(((((((((((---------(((..----((..((.(((.((........--------------...)).))---------).)).))...)-------)))))..)------))))))).)))).))))))).))...... ( -39.20, z-score = -3.12, R) >droGri2.scaffold_15126 799024 111 + 8399593 CUGCCAAGUCAUGGAGGCAAUCAUUUGGA---------CUCGU----CGCUGCUGCUCUAUCACUUUA--------------ACUGACAG---------UCGCUCGUCAG-------AGUCUCAG------GAUGAUUACAUUGAUGACGUUGCCACAUA ..((...(((((.((.(.(((((((((((---------(((..----.((.((((.((..........--------------...)))))---------).))......)-------)))))..)------))))))).).)).)))))...))...... ( -29.62, z-score = -0.08, R) >consensus UUGCCACGUCACGGAGGCAAUCAUUUGGGGCGC_CCCCCUCCU___GCAUAUCCUCUCAAAGUCCCCC____________CUGCUCCCAG________CCCUUUCGCUGACAGACACAGUCGUCG______CCUGAUUACUUUGAUGACGUUGCCACAUA ..((.((((((..((((.(((((...(((((.......)))))...............................................................(((.......)))..............))))).))))..)))))).))...... (-17.16 = -17.91 + 0.75)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:25 2011