
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,457,785 – 14,457,895 |
| Length | 110 |
| Max. P | 0.977896 |

| Location | 14,457,785 – 14,457,880 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 89.21 |
| Shannon entropy | 0.21205 |
| G+C content | 0.32808 |
| Mean single sequence MFE | -17.60 |
| Consensus MFE | -14.77 |
| Energy contribution | -14.69 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.84 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.98 |
| SVM RNA-class probability | 0.977896 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
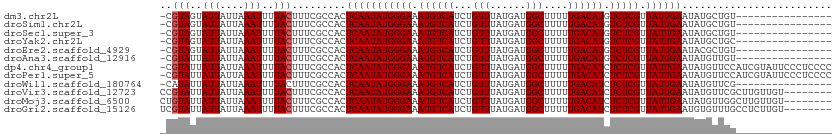
>dm3.chr2L 14457785 95 + 23011544 ----------------ACAGCAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG- ----------------...((.(((((((((..(.((.((((((...................)))))).)).).))))))))))).........................- ( -14.81, z-score = -1.88, R) >droSim1.chr2L 14206337 95 + 22036055 ----------------ACAGCAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG- ----------------...((.(((((((((..(.((.((((((...................)))))).)).).))))))))))).........................- ( -14.81, z-score = -1.88, R) >droSec1.super_3 716808 95 + 7220098 ----------------ACAGCAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG- ----------------...((.(((((((((..(.((.((((((...................)))))).)).).))))))))))).........................- ( -14.81, z-score = -1.88, R) >droYak2.chr2L 14564387 95 + 22324452 ----------------GCAGCAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG- ----------------...((.(((((((((..(.((.((((((...................)))))).)).).))))))))))).........................- ( -14.81, z-score = -1.64, R) >droEre2.scaffold_4929 6213024 95 + 26641161 ----------------ACAGCGUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG- ----------------....((((.........(.((.((((((...................)))))).)).).((((((((.((....))...))))))))....))))- ( -15.51, z-score = -1.73, R) >droAna3.scaffold_12916 14817623 95 - 16180835 ----------------ACAACAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG- ----------------......(((((((((..(.((.((((((...................)))))).)).).)))))))))((....))...................- ( -14.51, z-score = -2.12, R) >dp4.chr4_group1 1846149 111 - 5278887 GGGGAGGGAAUACGAUGGAACAUAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG- (((((((.....)(((((.((((.(((........))))))).......)))))..............)))))).((((((((.((....))...))))))))........- ( -22.40, z-score = -2.34, R) >droPer1.super_5 1787213 111 + 6813705 GGGGAGGGAAUACGAUGGAACAUAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG- (((((((.....)(((((.((((.(((........))))))).......)))))..............)))))).((((((((.((....))...))))))))........- ( -22.40, z-score = -2.34, R) >droWil1.scaffold_180764 1198090 95 - 3949147 ----------------CGAACAUAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUAUG- ----------------....((((((.......(.(((((((((...................))))))))).).((((((((.((....))...))))))))..))))))- ( -19.61, z-score = -3.72, R) >droVir3.scaffold_12723 3077018 104 - 5802038 --------ACAACAAGCGAACAUAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACGG --------........((....(((((((((..(.(((((((((...................))))))))).).)))))))))((....)).................)). ( -19.01, z-score = -2.95, R) >droMoj3.scaffold_6500 16386365 104 - 32352404 --------ACAACAAGCCAACAUAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACAG --------.......((((.....(((((((..(.(((((((((...................))))))))).).))))))))))).......................... ( -18.11, z-score = -3.54, R) >droGri2.scaffold_15126 783246 104 + 8399593 --------ACAAGAGGCAAACACAUUCAAUAACGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACGA --------..............(((((((((..(.(((((((((...................))))))))).).)))))))))((....)).................... ( -20.41, z-score = -3.54, R) >consensus ________________ACAACAUAUUCAAUAACGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG_ ......................(((((((((..(.((.((((((...................)))))).)).).)))))))))((....)).................... (-14.77 = -14.69 + -0.08)


| Location | 14,457,785 – 14,457,880 |
|---|---|
| Length | 95 |
| Sequences | 12 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 89.21 |
| Shannon entropy | 0.21205 |
| G+C content | 0.32808 |
| Mean single sequence MFE | -19.91 |
| Consensus MFE | -17.82 |
| Energy contribution | -17.90 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.86 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.939119 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14457785 95 - 23011544 -CGUAGUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUAUGCUGU---------------- -.((((.(((....))).))))....((...(((((((((((.((((((...(((......)))....)))))).))))).))))))....))...---------------- ( -19.70, z-score = -1.61, R) >droSim1.chr2L 14206337 95 - 22036055 -CGUAGUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUAUGCUGU---------------- -.((((.(((....))).))))....((...(((((((((((.((((((...(((......)))....)))))).))))).))))))....))...---------------- ( -19.70, z-score = -1.61, R) >droSec1.super_3 716808 95 - 7220098 -CGUAGUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUAUGCUGU---------------- -.((((.(((....))).))))....((...(((((((((((.((((((...(((......)))....)))))).))))).))))))....))...---------------- ( -19.70, z-score = -1.61, R) >droYak2.chr2L 14564387 95 - 22324452 -CGUAGUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUAUGCUGC---------------- -.(((((((......................(((((((((((.((((((...(((......)))....)))))).))))).))))))..)))))))---------------- ( -20.75, z-score = -1.93, R) >droEre2.scaffold_4929 6213024 95 - 26641161 -CGUAGUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUACGCUGU---------------- -.((((.(((....))).))))....((...(((((((((((.((((((...(((......)))....)))))).))))).))))))....))...---------------- ( -19.70, z-score = -1.56, R) >droAna3.scaffold_12916 14817623 95 + 16180835 -CGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUGUCUCGUUAUUGAAUAUGUUGU---------------- -.(((..(((....)))..))).........(((((((((((.((((((...(((......)))....)))))).))))).)))))).........---------------- ( -18.90, z-score = -1.67, R) >dp4.chr4_group1 1846149 111 + 5278887 -CGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUCCAUCGUAUUCCCUCCCC -.(((..(((....)))..))).........(((((((((((.((((((...(((......)))....)))))).))))).))))))(((((......)))))......... ( -20.10, z-score = -1.99, R) >droPer1.super_5 1787213 111 - 6813705 -CGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUCCAUCGUAUUCCCUCCCC -.(((..(((....)))..))).........(((((((((((.((((((...(((......)))....)))))).))))).))))))(((((......)))))......... ( -20.10, z-score = -1.99, R) >droWil1.scaffold_180764 1198090 95 + 3949147 -CAUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUCG---------------- -..............................(((((((((((.((((((...(((......)))....)))))).))))).)))))).........---------------- ( -17.80, z-score = -2.06, R) >droVir3.scaffold_12723 3077018 104 + 5802038 CCGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUCGCUUGUUGU-------- ..(((..(((....)))..))).........(((((((((((.((((((...(((......)))....)))))).))))).)))))).................-------- ( -18.90, z-score = -1.34, R) >droMoj3.scaffold_6500 16386365 104 + 32352404 CUGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUGGCUUGUUGU-------- ..(((..(((....)))..)))....((((((((((((((((.((((((...(((......)))....)))))).))))).))))))....).)))).......-------- ( -22.80, z-score = -2.67, R) >droGri2.scaffold_15126 783246 104 - 8399593 UCGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUGUGUUUGCCUCUUGU-------- ..(((..(((....)))..)))....((((.(((((((((((.((((((...(((......)))....)))))).))))).)))))).)).))...........-------- ( -20.80, z-score = -2.25, R) >consensus _CGUAUUAUUAUUAAAUUUUACUUUCGCCACUCAAUAUGGGAAAUGUCAUCUGUUUAUGAUGGCUUUUUGACAUCUCUCGUUAUUGAAUAUGUUGU________________ ..(((..(((....)))..))).........(((((((((((.((((((...(((......)))....)))))).))))).))))))......................... (-17.82 = -17.90 + 0.08)

| Location | 14,457,801 – 14,457,895 |
|---|---|
| Length | 94 |
| Sequences | 11 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.20 |
| Shannon entropy | 0.29386 |
| G+C content | 0.40030 |
| Mean single sequence MFE | -19.54 |
| Consensus MFE | -15.52 |
| Energy contribution | -15.24 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.11 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.79 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.855299 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14457801 94 + 23011544 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG----CCAUCGACGACGGGC---------------------- .......((((.....(.((((.......)))).)..........((((.(((((..((((...........)))).))----))))))).))))...---------------------- ( -16.70, z-score = -0.83, R) >droSim1.chr2L 14206353 94 + 22036055 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG----CCAUCGACGACGGGC---------------------- .......((((.....(.((((.......)))).)..........((((.(((((..((((...........)))).))----))))))).))))...---------------------- ( -16.70, z-score = -0.83, R) >droSec1.super_3 716824 94 + 7220098 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG----CCAUCGACGACGGGC---------------------- .......((((.....(.((((.......)))).)..........((((.(((((..((((...........)))).))----))))))).))))...---------------------- ( -16.70, z-score = -0.83, R) >droYak2.chr2L 14564403 94 + 22324452 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG----CCAGCGACGACGGGC---------------------- ......((((((...................))))))..(((.((((((((.((....))...))))))))......((----....)).....))).---------------------- ( -16.61, z-score = -0.77, R) >droEre2.scaffold_4929 6213040 94 + 26641161 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUACUACG----CCAACGACGAUGGGC---------------------- ......((((((...................))))))..(((((......(((((..((((...........)))).))----)))......))))).---------------------- ( -18.61, z-score = -1.73, R) >droAna3.scaffold_12916 14817639 93 - 16180835 CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG----ACAUCGGCGACGGC----------------------- ......(((((.....(.((((.......)))).)........((((((((.((....))...)))))))).......)----))))((....))..----------------------- ( -18.30, z-score = -1.61, R) >dp4.chr4_group1 1846181 102 - 5278887 CGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG----ACAACGACAGCAGCGGCAUCGGC-------------- .(.(((((((((...................))))))))).).((((((((.((....))...))))))))........----....(((..((....)).)))..-------------- ( -20.31, z-score = -1.77, R) >droPer1.super_5 1787245 108 + 6813705 CGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG----ACAACGACAGCAGCGGCAUCGACAUCGGC-------- (((..((((((.....(.((((.......)))).)........((((((((.((....))...))))))))........----..............))))))....)))..-------- ( -22.70, z-score = -2.05, R) >droWil1.scaffold_180764 1198106 92 - 3949147 CGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUAUG----UCCACGACAACGA------------------------ .(.(((((((((...................))))))))).).((((((((.((....))...))))))))......((----((...))))....------------------------ ( -19.01, z-score = -3.04, R) >droVir3.scaffold_12723 3077042 120 - 5802038 CGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACGGAAUACACCGACGGCGACAGCAACAGCGACAGCGACAGCAA .......((((.....(((.((((......)))).........((((((((.((....))...))))))))......(((......)))..)))....((....))))))((....)).. ( -25.90, z-score = -2.98, R) >droMoj3.scaffold_6500 16386389 114 - 32352404 CGAGAGAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACAGAAUACACCGACGGCGACGGCGACAGCGACAGCGA------ .(.(((((((((...................))))))))).).((((((((.((....))...))))))))................((.((....)).))...((....))..------ ( -23.41, z-score = -2.05, R) >consensus CGAGACAUGUCAAAAAGCCAUCAUAAACAGAUGACAUUUCCCAUAUUGAGUGGCGAAAGUAAAAUUUAAUAAUAAUACG____ACAACGACGACGGCC______________________ .......((((.....(.((((.......)))).)........((((((((.((....))...)))))))).................))))............................ (-15.52 = -15.24 + -0.28)



Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:22 2011