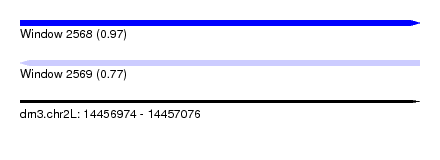
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,456,974 – 14,457,076 |
| Length | 102 |
| Max. P | 0.966520 |

| Location | 14,456,974 – 14,457,076 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 83.99 |
| Shannon entropy | 0.33686 |
| G+C content | 0.42191 |
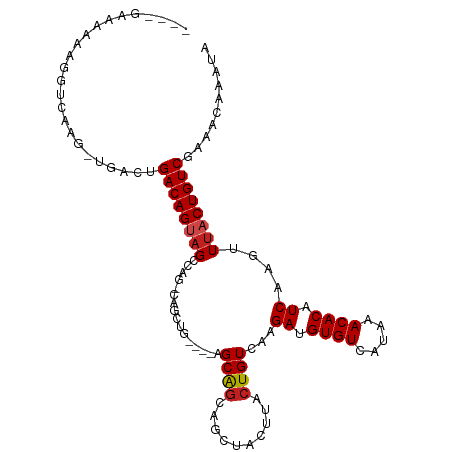
| Mean single sequence MFE | -30.71 |
| Consensus MFE | -18.64 |
| Energy contribution | -18.70 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.58 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.77 |
| SVM RNA-class probability | 0.966520 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
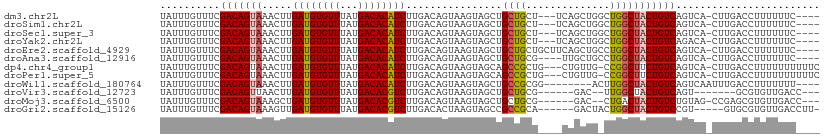
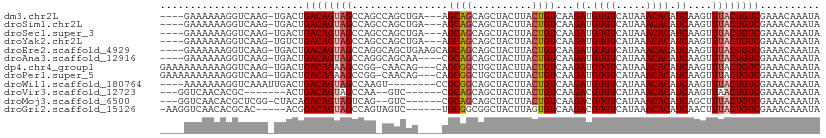
>dm3.chr2L 14456974 102 + 23011544 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCU---UCAGCUGGCUGGCUACUGUCAGUCA-CUUGACCUUUUUUC---- ..........(((.........((((((((...)))))))).((((((((..(((((((((..---.)))).)))))..))))))))))).-..............---- ( -33.40, z-score = -3.27, R) >droSim1.chr2L 14205553 102 + 22036055 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCU---UCAGCUGGCUGGCUACUGUCAGUCA-CUUGACCUUUUUUC---- ..........(((.........((((((((...)))))))).((((((((..(((((((((..---.)))).)))))..))))))))))).-..............---- ( -33.40, z-score = -3.27, R) >droSec1.super_3 716010 102 + 7220098 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCU---UCAGCUGGCUGGCUACUGUCAGUCA-CUUGACCUUUUUUC---- ..........(((.........((((((((...)))))))).((((((((..(((((((((..---.)))).)))))..))))))))))).-..............---- ( -33.40, z-score = -3.27, R) >droYak2.chr2L 14563560 102 + 22324452 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCU---UCAGCUGGCUGGCUACUGUCAGACA-CUUGACCUUUUUUC---- ....(((((.(((((((..(..((((((((...))))))))..).((((...((((((.....---.))))))))))..))))))))))))-..............---- ( -32.10, z-score = -2.97, R) >droEre2.scaffold_4929 6212249 105 + 26641161 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCUGCUUCAGCUGCCUGGCUACUGUCAGUCA-CUUGACCUUUUUUC---- ..........(((((((..(..((((((((...))))))))....(((((((((((....)))))))..))))...)..))))))).(((.-...)))........---- ( -33.30, z-score = -3.09, R) >droAna3.scaffold_12916 14816849 101 - 16180835 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCG----UUGCUGCCUGGCUACUGUCAGUCA-CUUGACCUUUUUUC---- ..........(((((((..(..((((((((...))))))))..).(((..((((((......)----)))))..)))..))))))).(((.-...)))........---- ( -30.90, z-score = -2.90, R) >dp4.chr4_group1 1845315 105 - 5278887 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCAGCCGCUG---CUGUUG-CCGGCUUCUGUCAGUCA-CUUGACCUUUUUUUUUUC ..........(((((....(..((((((((...))))))))..)..(((..((((((((....)---))))))-)..))).))))).(((.-...)))............ ( -30.90, z-score = -3.07, R) >droPer1.super_5 1786359 105 + 6813705 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCAGCCGCUG---CUGUUG-CCGGCUUCUGUCAGUCA-CUUGACCUUUUUUUUUUC ..........(((((....(..((((((((...))))))))..)..(((..((((((((....)---))))))-)..))).))))).(((.-...)))............ ( -30.90, z-score = -3.07, R) >droWil1.scaffold_180764 2749417 98 + 3949147 UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCCGCGG--------ACUUGGCUACUGUCAGUCAAUUUGACCUUUUUUU---- ..........(((((((..(..((((((((...))))))))..)....(((((.((....))..--------)))))..))))))).(((.....)))........---- ( -25.80, z-score = -2.00, R) >droVir3.scaffold_12723 3076292 92 - 5802038 UAUUUGUUUCGACAGUUAACUUGAUGUGUUUAUGACACGUCUUGACAGUAAGUAGCUGCUGCG------GAC--UUGGCUACUGUCAGU-------GCGUGUUGACC--- ........((((((((......((((((((...))))))))((((((((((((.((....)).------.))--)....))))))))).-------)).))))))..--- ( -27.10, z-score = -1.19, R) >droMoj3.scaffold_6500 16385530 98 - 32352404 UAUUUGUUUCGACAGUAAAGCUGAUGUGUUUAUGACACGUCUUGACAGUAAGUAGCUGCUGCG------GAC--CUGACUACUGUCUGUAG-CCGAGCGUGUUGACC--- ........((((((((..((((((((((((...))))))))...((.....))))))((((((------(((--.........))))))))-)...)).))))))..--- ( -29.20, z-score = -1.47, R) >droGri2.scaffold_15126 782098 98 + 8399593 UAUUUGUUUCGACAGUAAAGUUGAUGUGUUUAUGACACGUCUUGACACUAAGUAGCCGCCGCA------GACUACUGGCUACUGUCCGU-----GUGCGUGUUGACCUU- ........((((((((......((((((((...))))))))...((((..((((((((.....------......))))))))....))-----)))).))))))....- ( -28.10, z-score = -1.38, R) >consensus UAUUUGUUUCGACAGUAAACUUGAUGUGUUUAUGACACAUCUUGACAGUAAGUAGCUGCUGCU____CAGCUG_CUGGCUACUGUCAGUCA_CUUGACCUUUUUUC____ ..........(((((((.....((((((((...)))))))).............((....)).................)))))))........................ (-18.64 = -18.70 + 0.06)
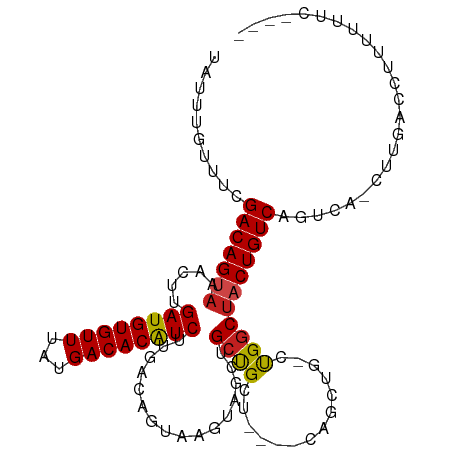
| Location | 14,456,974 – 14,457,076 |
|---|---|
| Length | 102 |
| Sequences | 12 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 83.99 |
| Shannon entropy | 0.33686 |
| G+C content | 0.42191 |
| Mean single sequence MFE | -27.19 |
| Consensus MFE | -14.36 |
| Energy contribution | -14.47 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.65 |
| SVM RNA-class probability | 0.773831 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14456974 102 - 23011544 ----GAAAAAAGGUCAAG-UGACUGACAGUAGCCAGCCAGCUGA---AGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((...-.))))((((((((..(((..((((.---..)))).)))..))))))))..(((((((.....)))))))..((((.......))))..... ( -30.00, z-score = -3.00, R) >droSim1.chr2L 14205553 102 - 22036055 ----GAAAAAAGGUCAAG-UGACUGACAGUAGCCAGCCAGCUGA---AGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((...-.))))((((((((..(((..((((.---..)))).)))..))))))))..(((((((.....)))))))..((((.......))))..... ( -30.00, z-score = -3.00, R) >droSec1.super_3 716010 102 - 7220098 ----GAAAAAAGGUCAAG-UGACUGACAGUAGCCAGCCAGCUGA---AGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((...-.))))((((((((..(((..((((.---..)))).)))..))))))))..(((((((.....)))))))..((((.......))))..... ( -30.00, z-score = -3.00, R) >droYak2.chr2L 14563560 102 - 22324452 ----GAAAAAAGGUCAAG-UGUCUGACAGUAGCCAGCCAGCUGA---AGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----........(.((.(-((..(((((((((..(((..((((.---..)))).)))..))))))))).(((((((.....))))))).....))))).).......... ( -28.50, z-score = -2.52, R) >droEre2.scaffold_4929 6212249 105 - 26641161 ----GAAAAAAGGUCAAG-UGACUGACAGUAGCCAGGCAGCUGAAGCAGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((...-.))))((((((((.(.(((((...(((.(((....))).))).)))))..(((((((.....)))))))..).)))))))).......... ( -33.90, z-score = -3.46, R) >droAna3.scaffold_12916 14816849 101 + 16180835 ----GAAAAAAGGUCAAG-UGACUGACAGUAGCCAGGCAGCAA----CGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((...-.))))((((((((.(.(((((...----.((....))......)))))..(((((((.....)))))))..).)))))))).......... ( -28.70, z-score = -2.75, R) >dp4.chr4_group1 1845315 105 + 5278887 GAAAAAAAAAAGGUCAAG-UGACUGACAGAAGCCGG-CAACAG---CAGCGGCUGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ...........((((...-.))))(((((((((.((-((..((---.(((....))).))...))))..(((((((.....)))))))..)))).))))).......... ( -27.90, z-score = -2.41, R) >droPer1.super_5 1786359 105 - 6813705 GAAAAAAAAAAGGUCAAG-UGACUGACAGAAGCCGG-CAACAG---CAGCGGCUGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ...........((((...-.))))(((((((((.((-((..((---.(((....))).))...))))..(((((((.....)))))))..)))).))))).......... ( -27.90, z-score = -2.41, R) >droWil1.scaffold_180764 2749417 98 - 3949147 ----AAAAAAAGGUCAAAUUGACUGACAGUAGCCAAGU--------CCGCGGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ----.......((((.....))))((((((((.(....--------....(((((.......)))))..(((((((.....)))))))..).)))))))).......... ( -28.20, z-score = -3.84, R) >droVir3.scaffold_12723 3076292 92 + 5802038 ---GGUCAACACGC-------ACUGACAGUAGCCAA--GUC------CGCAGCAGCUACUUACUGUCAAGACGUGUCAUAAACACAUCAAGUUAACUGUCGAAACAAAUA ---(((.(((....-------..(((((((((...(--((.------.......)))..))))))))).((.((((.....)))).))..))).)))............. ( -18.40, z-score = -0.49, R) >droMoj3.scaffold_6500 16385530 98 + 32352404 ---GGUCAACACGCUCGG-CUACAGACAGUAGUCAG--GUC------CGCAGCAGCUACUUACUGUCAAGACGUGUCAUAAACACAUCAGCUUUACUGUCGAAACAAAUA ---...........((((-(....((((((((...(--((.------.......)))..))))))))..((.((((.....)))).)).........)))))........ ( -20.40, z-score = -0.10, R) >droGri2.scaffold_15126 782098 98 - 8399593 -AAGGUCAACACGCAC-----ACGGACAGUAGCCAGUAGUC------UGCGGCGGCUACUUAGUGUCAAGACGUGUCAUAAACACAUCAACUUUACUGUCGAAACAAAUA -...............-----...((((((((..(((.(((------(..((((.((....)))))).))))((((.....))))....))))))))))).......... ( -22.40, z-score = -0.36, R) >consensus ____GAAAAAAGGUCAAG_UGACUGACAGUAGCCAG_CAGCUG____AGCAGCAGCUACUUACUGUCAAGAUGUGUCAUAAACACAUCAAGUUUACUGUCGAAACAAAUA ........................((((((((................((((..........))))...((.((((.....)))).))....)))))))).......... (-14.36 = -14.47 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:20 2011