| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,450,210 – 14,450,301 |
| Length | 91 |
| Max. P | 0.602088 |

| Location | 14,450,210 – 14,450,301 |
|---|---|
| Length | 91 |
| Sequences | 9 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 71.91 |
| Shannon entropy | 0.52220 |
| G+C content | 0.37172 |
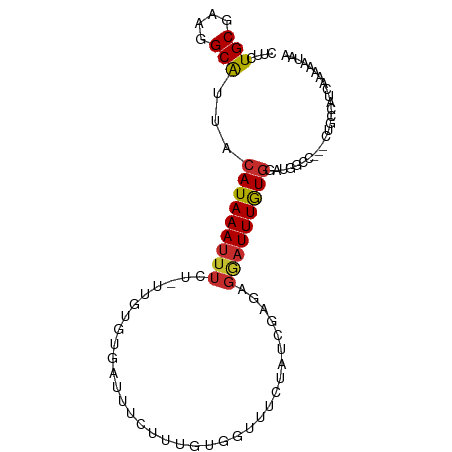
| Mean single sequence MFE | -22.77 |
| Consensus MFE | -6.75 |
| Energy contribution | -6.35 |
| Covariance contribution | -0.40 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.30 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.602088 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
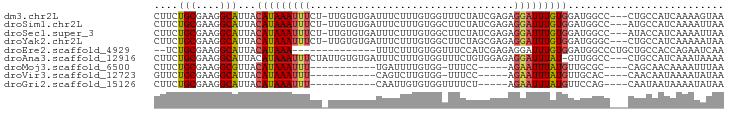
>dm3.chr2L 14450210 91 - 23011544 CUUCUGCGAAGGCAUUACAUAAAUUUCU-UUGUGUGAUUUCUUUGUGGUUUCUAUCGAGAGGAUUUGUGGAUGGCC---CUGCCAUCAAAAGUAA ((((((((((((.((((((((((....)-))))))))).)))))))(((....))).)))))((((...((((((.---..))))))..)))).. ( -28.40, z-score = -3.16, R) >droSim1.chr2L 14198929 91 - 22036055 CUUCUGCGAAGGCAUUACAUAAAUUUCU-UUGUGUGAUUUCUUUGUGGCUUCUAUCGAGAGGAUUUGUGGAUGGCC---AUGCCAUCAAAAUUAA ...(..((((((.((((((((((....)-))))))))).))))))..)(((((....))))).......((((((.---..))))))........ ( -28.80, z-score = -3.32, R) >droSec1.super_3 709335 91 - 7220098 CUUCUGCGAAGGCAUUACAUAAAUUUCU-UUGUGUGAUUUCUUUGUGGCUUCUAUCGAGAGGAUUUGUGGAUGGCC---AUACCAUCAAAAUUAA ...(..((((((.((((((((((....)-))))))))).))))))..)(((((....))))).......(((((..---...)))))........ ( -26.60, z-score = -3.02, R) >droYak2.chr2L 14556624 91 - 22324452 CUUCUGCGAAGGCAUUACAUAAAUUUCU-UUGUGUGAUUUCUUUGUGGCUUCUAGCGAGAGGAUUUGUGGAUGGGC---CUGCCAUCAAAAAUAA ...(..((((((.((((((((((....)-))))))))).))))))..)(((((....)))))((((...(((((..---...)))))..)))).. ( -27.90, z-score = -2.51, R) >droEre2.scaffold_4929 6205348 79 - 26641161 --UCUGCGAAGGCAUUACAUAAA--------------UUUCUUUGUGGUUUCCAUCGAGAGGAUUUGUGGAUGGCCCUGCUGCCACCAGAAUCAA --..(((....))).........--------------.(((((.((((...)))).)))))(((((.(((.((((......)))))))))))).. ( -22.70, z-score = -1.18, R) >droAna3.scaffold_12916 14810381 91 + 16180835 CUUCUGCGAAGGCAUUACAUAAAUUUCUAUUGUGUGAUUUCUUUGUGGUUUCUGUGGAGAGGAUUUAU-GUUGGCC---CUGCCAUCAAAUAAAA ...(..((((((.(((((((((.......))))))))).))))))..)(((((....))))).(((((-..((((.---..))))....))))). ( -22.50, z-score = -1.59, R) >droMoj3.scaffold_6500 16374976 74 + 32352404 CUUCUGCGAAGGCGUUACAUAAAUUU-----------UGAUUUUGUGG-UUUCC-----AGAAUUUAUGUUGCGC----CAGCAACAAAAUUUAA ....(((...(((((.((((((((((-----------((.........-....)-----))))))))))).))))----).)))........... ( -23.42, z-score = -4.07, R) >droVir3.scaffold_12723 3069158 74 + 5802038 GUUCUGCGAAGGCAUUACAUAAAUUU-----------CAGUCUUGUGG-UUUCC-----AGAAUUUAUGUUGCAC----CAACAAUAAAAUAUAA ((((((.((((.(((.((........-----------..))...))).-)))))-----)))))...(((((...----)))))........... ( -12.40, z-score = -0.83, R) >droGri2.scaffold_15126 772874 75 - 8399593 CUUCUGCGAAGGCAUUACAUAAAUUU-----------CAAUUGUGUGGUUUUCU-----AGAAUUUAUGUUCCAG----CAAUAAUAAAAUAUAA ....(((((((((..(((((((....-----------...))))))))))))).-----.((((....))))..)----)).............. ( -12.20, z-score = -1.00, R) >consensus CUUCUGCGAAGGCAUUACAUAAAUUUCU_UUGUGUGAUUUCUUUGUGGUUUCUAUCGAGAGGAUUUGUGGAUGGCC___CUGCCAUCAAAAAUAA ....(((....)))...(((((((((..............((....))............))))))))).......................... ( -6.75 = -6.35 + -0.40)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:18 2011