| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,437,865 – 14,437,963 |
| Length | 98 |
| Max. P | 0.524213 |

| Location | 14,437,865 – 14,437,963 |
|---|---|
| Length | 98 |
| Sequences | 8 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 84.47 |
| Shannon entropy | 0.28595 |
| G+C content | 0.42622 |
| Mean single sequence MFE | -22.69 |
| Consensus MFE | -15.03 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.524213 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
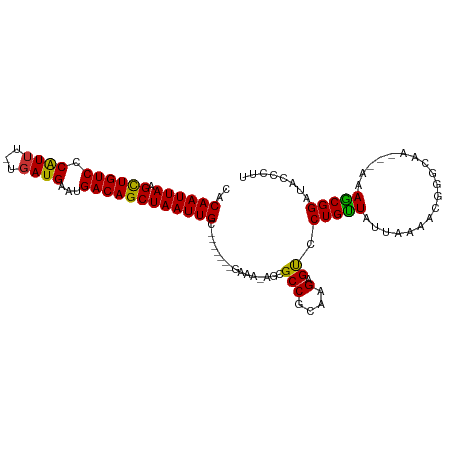
>dm3.chr2L 14437865 98 - 23011544 CACAAUUAAGCUGUCCCAUUU-UGAUGAAUGACAGCUAAUUGC-------GAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAAAAAAAAACGGAUACCCUU ........(((((((.(((..-..)))...)))))))..((((-------(...-.....)))))..(.((((((.....)))))))..........((....)).. ( -22.50, z-score = -1.85, R) >droEre2.scaffold_4929 6192085 95 - 26641161 CACAAUUAAGCUGUCCCAUUU-UGAUGAAUGACAGCUAAUUGC-------GAAA-AGCGCCUGGAGAGUCCUGUUAUUAAAGCGGGCAA---AAAGCGGAUACCCUU .........(((.((((.(((-((((((..(((..(((..(((-------....-.)))..)))...)))...))))))))).))).).---..)))((....)).. ( -22.10, z-score = -0.51, R) >droYak2.chr2L 14542725 95 - 22324452 CACAAUUAAGCUGUCCCGUUU-UGAUGAAUGACAGCUAAUUGC-------GAAA-AGCGCCUGCAGAGUCCUGUUAUUAAAACGGGCAA---AAAGCGGAUACCCUU .........(((.((((((((-((((((..(((.((.....))-------....-.((....))...)))...))))))))))))).).---..)))((....)).. ( -26.90, z-score = -2.45, R) >droSec1.super_3 697052 95 - 7220098 CACAAUUAAGCUGUCCCAUGU-UGAUGAAUGACAGCUAAUUGC-------GAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAA---AAAGCGGAUACCCUU (.((((((.((((((.(((..-..)))...)))))))))))).-------)...-....((((......((((((.....))))))...---...))))........ ( -26.40, z-score = -2.47, R) >droSim1.chr2L 14186616 95 - 22036055 CACAAUUAAGCUGUCCCAUUU-UGAUGAAUGACAGCUAAUUGC-------GAAA-AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAA---AAAGCGGAUACCCUU (.((((((.((((((.(((..-..)))...)))))))))))).-------)...-....((((......((((((.....))))))...---...))))........ ( -26.20, z-score = -2.59, R) >droPer1.super_5 1761645 92 - 6813705 -CCAAUUAAGUUGUCACAUUUCUGAUGAAUGACAGCUAAUUGCACAAA--AAAAAAGCGCCCCAAGAGCACUGCUAUUAA------------GAAGCGGAUACCCUU -.((((((.(((((((...(((....))))))))))))))))......--......((.........)).(((((.....------------..)))))........ ( -19.00, z-score = -1.91, R) >dp4.chr4_group1 1820589 94 + 5278887 -CCAAUUAAGUUGUCACAUUUCUGAUGAAUGACAGCUAAUUGCACAAAAUAAAAAAGCGCCCCAAGAGCACUGCUAUUAA------------GAAGCGGAUACCCUU -.((((((.(((((((...(((....))))))))))))))))..............((.........)).(((((.....------------..)))))........ ( -19.00, z-score = -1.92, R) >droAna3.scaffold_12916 14798630 96 + 16180835 UCCAAUUAAGCUGUCCCAUUU-UGAUGAAUGACAGCUAAUUGC-------GAAAGAGCGCCUCGAGAGUCCUGUUAUUAAAGCCGGCAA---AAAGCGGAUACCCUU ((((((((.((((((.(((..-..)))...)))))))))))).-------))..(((...)))....((((.(((.....)))..((..---...))))))...... ( -19.40, z-score = -0.24, R) >consensus CACAAUUAAGCUGUCCCAUUU_UGAUGAAUGACAGCUAAUUGC_______GAAA_AGCGCCGCAAGAGUCCUGUUAUUAAAACGGGCAA___AAAGCGGAUACCCUU ..((((((.((((((.(((.....)))...))))))))))))................(((....).)).(((((...................)))))........ (-15.03 = -14.25 + -0.78)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:15 2011