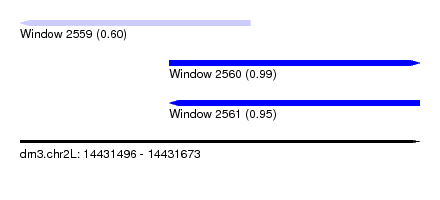
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,431,496 – 14,431,673 |
| Length | 177 |
| Max. P | 0.986233 |

| Location | 14,431,496 – 14,431,598 |
|---|---|
| Length | 102 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 72.19 |
| Shannon entropy | 0.54860 |
| G+C content | 0.53411 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -11.08 |
| Energy contribution | -12.60 |
| Covariance contribution | 1.52 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.21 |
| SVM RNA-class probability | 0.596242 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14431496 102 - 23011544 -AGCCAUUCUGC--CUUCCUGCCUGGCCGGCAGUUGGCCGGCACACAAAAUUGGUUUCUGUGUCU--GCUCGGCUGUUAAUCUC--UGUUGCAGAGAUUAGCGCCCCCA-------- -.((((..((((--(..((.....))..))))).)))).((((((.(((.....))).)))))).--....(((.(((((((((--((...))))))))))))))....-------- ( -42.10, z-score = -3.38, R) >droSec1.super_3 690712 99 - 7220098 -AGCCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCCGGCACACAAAAUUGGUUUCUGUGUCU--GCUCGGCUGUUAAUCUC--UGUUGCAGAGAUUAGCGCCCCCA-------- -((((.......--......(((.(((---(....)))))))(((((.((.....)).)))))..--....))))(((((((((--((...))))))))))).......-------- ( -36.30, z-score = -2.57, R) >droYak2.chr2L 14536246 99 - 22324452 -AGCCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCCGGCACACAAAAUUGGUUUCUGUGUCU--GCUCGGCUGUUAAUCUC--UGUUGCAGAGAUUAGCGCCCCCA-------- -((((.......--......(((.(((---(....)))))))(((((.((.....)).)))))..--....))))(((((((((--((...))))))))))).......-------- ( -36.30, z-score = -2.57, R) >droEre2.scaffold_4929 6184725 101 - 26641161 -AGCCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCUGGCACAGAAAAUUGGUUUCUGUGUCUGCGCCCGGUUGUUAAUCUC--UGUUGCAGAGAUUAGCGCCCCCA-------- -(((((..((((--(.........)).---))).)))))((((((((((.....)))))))))).......((.((((((((((--((...)))))))))))).))...-------- ( -40.90, z-score = -3.69, R) >droAna3.scaffold_12916 14791893 100 + 16180835 -AGCCAUUCUGC--UAGGCAGGCUGGC------CAGGCUGGAACCCAAAAUUGGUUGCCGAGUCUGUUCUCGUUUGUUAAUCUCUGUGUUGCAGAGAUUAGCGCCCCCA-------- -.((((..((((--...))))..))))------((((((((((((.......)))).)).)))))).....(..((((((((((((.....))))))))))))..)...-------- ( -40.30, z-score = -3.06, R) >dp4.chr4_group1 1811896 107 + 5278887 AGCCCAUUCUGUUGCCUGCUGCCUGG------GCUGACUGGCACACAAAAUUGGUUUCUGUGUCU--GUUCGGCAGUUAAUCUG--UGUUGCAGAGAUUAGCGCCCCCAACCCGCUC ((((((..(.((.....)).)..)))------)))(((.((((((.(((.....))).)))))).--))).(((.((((((((.--((...)).)))))))))))............ ( -32.30, z-score = -0.45, R) >droPer1.super_5 1752944 107 - 6813705 AGCCCAUUCUGUUGCCUGCUGCCUGG------GCUGACUGGCACACAAAAUUGGUUUCUGUGUCU--GUUCGGCUGUUAAUCUG--UGUUGCAGAGAUUAGCGCCCCCAACCCGCUC ((((((..(.((.....)).)..)))------)))(((.((((((.(((.....))).)))))).--))).(((.((((((((.--((...)).)))))))))))............ ( -32.30, z-score = -0.74, R) >droWil1.scaffold_180764 1238590 98 + 3949147 -------AGAGCAGAAUGCUGAAUGCCAUCCUUCUGACUGGCACACAAAAUUGGUUUCUGUGUCC--AUUCGCCUGUUAAUCUG--UGCAACAGAGAUUAGCGCCCCCA-------- -------((.((.(((((......((((((.....)).))))(((((.((.....)).))))).)--))))))))((((((((.--((...)).)))))))).......-------- ( -24.40, z-score = -0.58, R) >droGri2.scaffold_15126 748255 79 - 8399593 ----------------CACCGUUCACCAGUCACCGACCGGUCACACAAAAUUGGUUGC---------UCUCUG-UGUUAAUCUG--UGUAGCAGAGAUUAGCGCCCC---------- ----------------...........((....((((((((........)))))))).---------...))(-(((((((((.--((...)).))))))))))...---------- ( -19.50, z-score = -0.95, R) >consensus _AGCCAUUCUGC__CUUCCUGCCUGGC___CAGCUGGCUGGCACACAAAAUUGGUUUCUGUGUCU__GCUCGGCUGUUAAUCUC__UGUUGCAGAGAUUAGCGCCCCCA________ .......................................((((((.(((.....))).)))))).......(((.((((((((...........)))))))))))............ (-11.08 = -12.60 + 1.52)

| Location | 14,431,562 – 14,431,673 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 79.77 |
| Shannon entropy | 0.41725 |
| G+C content | 0.49306 |
| Mean single sequence MFE | -37.42 |
| Consensus MFE | -22.98 |
| Energy contribution | -22.50 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.44 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986233 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14431562 111 + 23011544 GGCCAACUGCCGGCCAGGCAGGAAG--GCAGAAUGG-CUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAAA-UUUACGGCUUUCUGGGUAAUUUGACACUUCG-GACAGAAUAAA (((((.(((((..((.....))..)--))))..)))-)).(((..((((((.(((.(.((((((((((....-.....)))))))))).)...))).))).)))-..)))...... ( -43.90, z-score = -3.23, R) >droSec1.super_3 690778 108 + 7220098 GGCCAACUG---GCCAGGCAGGAAG--GCAGAAUGG-CUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAAA-UUUACGGCUUUCUGGGUAAUUUGACACUUCG-GACAGAAUAAA ((((((..(---((((.((......--))....)))-)).))))))(((((.(((.(.((((((((((....-.....)))))))))).)...))).)))))..-........... ( -38.40, z-score = -2.32, R) >droYak2.chr2L 14536312 108 + 22324452 GGCCAACUG---GCCAGGCAGGAAG--GCAGAAUGG-CUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAAA-UUUACGGCUUUCUGGGUAAUUUGACACUUCG-GACAGAAUAAA ((((((..(---((((.((......--))....)))-)).))))))(((((.(((.(.((((((((((....-.....)))))))))).)...))).)))))..-........... ( -38.40, z-score = -2.32, R) >droEre2.scaffold_4929 6184793 108 + 26641161 AGCCAACUG---GCCAGGCAGGAAG--GCAGAAUGG-CUCUUGGUCGAGUGACAGCAGCCAGAAGGCCUAAA-UUUACGGCCUUCUGGGUAAUUUGACACUUUG-GACAGAAUGAA ......(((---.((((((((((.(--.(.....).-))))))(((((((.((.....((((((((((....-.....)))))))))))).)))))))..))))-).)))...... ( -38.10, z-score = -1.79, R) >droAna3.scaffold_12916 14791963 105 - 16180835 ---AGCCUG---GCCAGCCUGCCUA--GCAGAAUGG-CUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAA-UUUACGGCUUUCUGGGUAAUUUGACACUUUG-GACAGAAUAAU ---...(((---((((..((((...--))))..)))-)(((.((((((((.((.....((((((((((....-.....)))))))))))).))))))).)...)-)))))...... ( -37.50, z-score = -2.33, R) >dp4.chr4_group1 1811970 110 - 5278887 ---AGUCAG---CCCAGGCAGCAGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUUUUCGACAGAAUAUA ---.(((((---((((..(....(....).)..))))))....(((((((.((.....((((((((((..........)))))))))))).))))))).......)))........ ( -37.40, z-score = -3.03, R) >droPer1.super_5 1753018 110 + 6813705 ---AGUCAG---CCCAGGCAGCAGGCAACAGAAUGGGCUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAAAUUUACGGCUUUCUGGGUAAUUUGACACUUUUCGACAGAAUAUA ---.(((((---((((..(....(....).)..))))))....(((((((.((.....((((((((((..........)))))))))))).))))))).......)))........ ( -37.40, z-score = -3.03, R) >droWil1.scaffold_180764 1238656 108 - 3949147 -------AGUCAGAAGGAUGGCAUUCAGCAUUCUGCUCUCUUGGUCAAAUGACAGCAACCAGAAGGCCUAAA-UUUACGGCUUUCUGGGUAAUUUGACACUUUCGGACAGAAUAAA -------.(((.(((((....((...(((.....)))....))(((((((.((.....((((((((((....-.....)))))))))))).))))))).))))).)))........ ( -34.00, z-score = -2.98, R) >droGri2.scaffold_15126 748311 95 + 8399593 ---------------CGGUCGGUGACUGGUGAACGG-UGCUCGGCCAAAUGACAGCAGCCAGAAGGCCCGAA-UUUACGGCUUUCUGGGUAAUUUGACACUCUG-GACAGCAG--- ---------------.(((((((.((((.....)))-))).)))))......(((.((((((((((((....-.....))))))))))((......)).)))))-........--- ( -31.70, z-score = -0.90, R) >consensus ___CAACUG___GCCAGGCAGGAAG__GCAGAAUGG_CUCUUGGUCAAAUGACAGCAGCCAGAAGGCCUAAA_UUUACGGCUUUCUGGGUAAUUUGACACUUUG_GACAGAAUAAA ...........................................(((((((.((.....((((((((((..........)))))))))))).))))))).................. (-22.98 = -22.50 + -0.48)

| Location | 14,431,562 – 14,431,673 |
|---|---|
| Length | 111 |
| Sequences | 9 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 79.77 |
| Shannon entropy | 0.41725 |
| G+C content | 0.49306 |
| Mean single sequence MFE | -32.36 |
| Consensus MFE | -18.98 |
| Energy contribution | -18.34 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.13 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.58 |
| SVM RNA-class probability | 0.951860 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14431562 111 - 23011544 UUUAUUCUGUC-CGAAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAG-CCAUUCUGC--CUUCCUGCCUGGCCGGCAGUUGGCC .....((((..-(((.(((.((...((.((((((.(((.....-....))).)))))).)).)).))))))..).)))(-(((..((((--(..((.....))..))))).)))). ( -38.70, z-score = -3.26, R) >droSec1.super_3 690778 108 - 7220098 UUUAUUCUGUC-CGAAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAG-CCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCC .....((((..-(((.(((.((...((.((((((.(((.....-....))).)))))).)).)).))))))..).)))(-(((..((((--(.........)).---))).)))). ( -32.00, z-score = -2.20, R) >droYak2.chr2L 14536312 108 - 22324452 UUUAUUCUGUC-CGAAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAG-CCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCC .....((((..-(((.(((.((...((.((((((.(((.....-....))).)))))).)).)).))))))..).)))(-(((..((((--(.........)).---))).)))). ( -32.00, z-score = -2.20, R) >droEre2.scaffold_4929 6184793 108 - 26641161 UUCAUUCUGUC-CAAAGUGUCAAAUUACCCAGAAGGCCGUAAA-UUUAGGCCUUCUGGCUGCUGUCACUCGACCAAGAG-CCAUUCUGC--CUUCCUGCCUGGC---CAGUUGGCU .....((((((-...((((.((...((.((((((((((.....-....)))))))))).)).)).)))).)))..)))(-(((..((((--(.........)).---))).)))). ( -37.50, z-score = -3.28, R) >droAna3.scaffold_12916 14791963 105 + 16180835 AUUAUUCUGUC-CAAAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAG-CCAUUCUGC--UAGGCAGGCUGGC---CAGGCU--- .....((((..-....(.(((((((.((((((((.(((.....-....))).)))))).....)).))))))))....(-(((..((((--...))))..))))---))))..--- ( -33.50, z-score = -1.39, R) >dp4.chr4_group1 1811970 110 + 5278887 UAUAUUCUGUCGAAAAGUGUCAAAUUACCCAGAAAGCCGUAAAUUUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGCCCAUUCUGUUGCCUGCUGCCUGGG---CUGACU--- ........(((.....(.(((((((.((((((((.(((..........))).)))))).....)).))))))))...((((((..(.((.....)).)..))))---))))).--- ( -33.00, z-score = -2.45, R) >droPer1.super_5 1753018 110 - 6813705 UAUAUUCUGUCGAAAAGUGUCAAAUUACCCAGAAAGCCGUAAAUUUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAGCCCAUUCUGUUGCCUGCUGCCUGGG---CUGACU--- ........(((.....(.(((((((.((((((((.(((..........))).)))))).....)).))))))))...((((((..(.((.....)).)..))))---))))).--- ( -33.00, z-score = -2.45, R) >droWil1.scaffold_180764 1238656 108 + 3949147 UUUAUUCUGUCCGAAAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUUAGGCCUUCUGGUUGCUGUCAUUUGACCAAGAGAGCAGAAUGCUGAAUGCCAUCCUUCUGACU------- ..(((((((..(....(.(((((((.((((((((.(((.....-....))).)))))).....)).))))))))....)..))))))).....................------- ( -28.40, z-score = -2.23, R) >droGri2.scaffold_15126 748311 95 - 8399593 ---CUGCUGUC-CAGAGUGUCAAAUUACCCAGAAAGCCGUAAA-UUCGGGCCUUCUGGCUGCUGUCAUUUGGCCGAGCA-CCGUUCACCAGUCACCGACCG--------------- ---..((((..-....(.(((((((.((((((((.(((.....-....))).)))))).....)).))))))))((((.-..))))..)))).........--------------- ( -23.10, z-score = 0.27, R) >consensus UUUAUUCUGUC_CAAAGUGUCAAAUUACCCAGAAAGCCGUAAA_UUUAGGCCUUCUGGCUGCUGUCAUUUGACCAAGAG_CCAUUCUGC__CUUCCUGCCUGGC___CAGUUG___ ....(((((((....((((.((...((.((((((.(((..........))).)))))).)).)).)))).)))..))))..................................... (-18.98 = -18.34 + -0.64)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:14 2011