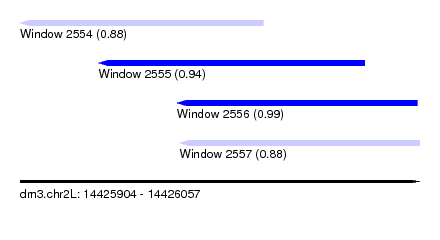
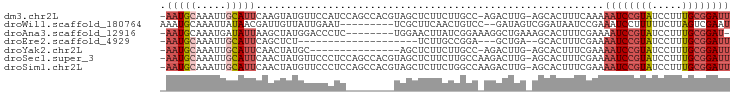
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,425,904 – 14,426,057 |
| Length | 153 |
| Max. P | 0.991497 |

| Location | 14,425,904 – 14,425,997 |
|---|---|
| Length | 93 |
| Sequences | 10 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 62.99 |
| Shannon entropy | 0.71148 |
| G+C content | 0.46272 |
| Mean single sequence MFE | -21.41 |
| Consensus MFE | -7.10 |
| Energy contribution | -7.02 |
| Covariance contribution | -0.08 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.33 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.880286 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
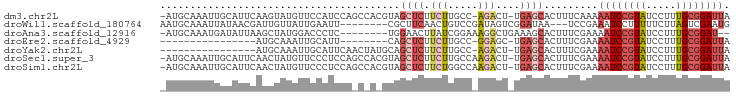
>dm3.chr2L 14425904 93 - 23011544 GCACUUUCAAAAAUCCGUAUCCUUUGCGGAUUACA-AAACAGA-------------ACCAAUAUC---CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG .(((.(((...((((((((.....))))))))...-.....((-------------..((((...---...)))))).....))).)))((((((...))))))...... ( -21.50, z-score = -2.15, R) >droGri2.scaffold_15126 740997 101 - 8399593 ----GCUUCCCGCGUCAACGGGUCGAGUCCUUUGGAUUACAUGCACAUC---CAAUUCCAUUGUC-AGCUCGUCGUC-GUUGAAGUUUCGCCUUUGAUGAAGGCGACACG ----((.....)).((((((((.(((((.(..((((.............---....))))..)..-.))))).).))-))))).((.((((((((...)))))))).)). ( -31.63, z-score = -1.77, R) >droVir3.scaffold_12723 3039635 93 + 5802038 ----GCUCCC----UCAAUGUGGCGAGU-CUUUGGAUUACAUACACAUU---CAAUUCCAUUGCC-AAC---UCAAC-GCUCGAGUUUCGCCUUUGAUGAAGGCGACACG ----.....(----((...(((..((((-(..((((.............---....))))..)..-.))---))..)-))..)))..((((((((...)))))))).... ( -22.13, z-score = -1.30, R) >droPer1.super_5 1741066 86 - 6813705 -----------UAUCCACUUUCACAAAUCCUUACACAAACAGA-------------ACGAAUAUCCAUCCCAUUGUCAAGUUGAAAGCGGCCUUUGAUGAAGGCGACACG -----------................................-------------.................((((..((.....)).((((((...)))))))))).. ( -10.20, z-score = 0.53, R) >dp4.chr4_group1 1800101 86 + 5278887 -----------UAUCCACUUUCACAAAUCCUUACACAAACAGA-------------ACGAAUAUCCAUCCCAUUGUCAAGUUGAAAGCGGCCUUUGAUGAAGGCGACACG -----------................................-------------.................((((..((.....)).((((((...)))))))))).. ( -10.20, z-score = 0.53, R) >droAna3.scaffold_12916 14786191 76 + 16180835 ------------AAGCACUUUCGAAAAUCCGUAUCCUUUGCGG-------------------AUC---CCCAUUGUCAACUCGAAUGCGGCCUUUGAUGAAGGCGACACG ------------..(((..(((((..(((((((.....)))))-------------------)).---......(....))))))))).((((((...))))))...... ( -19.60, z-score = -1.27, R) >droEre2.scaffold_4929 6179254 93 - 26641161 --------------GCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACCAAACAGAACCAAUAUC---CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG --------------.(((.(((((.((((((((.....))))))))........((..((((...---...))))))...))))).)))((((((...))))))...... ( -24.70, z-score = -3.01, R) >droYak2.chr2L 14530572 93 - 22324452 --------------GCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUC---CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG --------------.(((.(((((.((((((((.....))))))))........((..((((...---...))))))...))))).)))((((((...))))))...... ( -24.70, z-score = -2.98, R) >droSec1.super_3 685333 93 - 7220098 --------------GCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUC---CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG --------------.(((.(((((.((((((((.....))))))))........((..((((...---...))))))...))))).)))((((((...))))))...... ( -24.70, z-score = -2.98, R) >droSim1.chr2L 14174089 93 - 22036055 --------------GCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUC---CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG --------------.(((.(((((.((((((((.....))))))))........((..((((...---...))))))...))))).)))((((((...))))))...... ( -24.70, z-score = -2.98, R) >consensus ______________GCACUUUCGAAAAUCCGUAUCCUUACAGG___A_____CA__ACCAAUAUC___CCCAUUGUCAACUCGAAUGUGGCCUUUGAUGAAGGCGACACG .........................................................................((((....((....))((((((...)))))))))).. ( -7.10 = -7.02 + -0.08)

| Location | 14,425,934 – 14,426,036 |
|---|---|
| Length | 102 |
| Sequences | 5 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 85.77 |
| Shannon entropy | 0.23887 |
| G+C content | 0.45403 |
| Mean single sequence MFE | -18.04 |
| Consensus MFE | -16.06 |
| Energy contribution | -15.90 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.80 |
| Structure conservation index | 0.89 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.940026 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14425934 102 - 23011544 UGUUCCAUCCAGCCACGUAGCUCUUCUUGCC-AGACUUGAGCACUUUCAAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAAC .((((...........((.((((.(((....-)))...)))))).......((((((((.....))))))))........))))................... ( -18.50, z-score = -2.08, R) >droEre2.scaffold_4929 6179284 83 - 26641161 -------------------GCUCUUCUUGCC-GGAGCUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACCAAACAGAACCAAUAUCCCCAUUGUCAAC -------------------((((..(((...-.)))..))))......((.((((((((.....))))))))............((((......))))))... ( -17.20, z-score = -1.89, R) >droYak2.chr2L 14530602 87 - 22324452 ---------------UGCAGCUCUUCUUGCC-AGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAAC ---------------.(((((((.(((....-)))...)))).........((((((((.....))))))))........................))).... ( -16.00, z-score = -1.61, R) >droSec1.super_3 685363 103 - 7220098 UGUUCCCUCCAGCCACGUAGCUCUUCUUGCCAAGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAAC .((((...........((.((((.(((.....)))...)))))).......((((((((.....))))))))........))))................... ( -18.70, z-score = -1.87, R) >droSim1.chr2L 14174119 103 - 22036055 UGUUCCCUCCAGCCACGUAGCUCUUCUGGCCAAGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAAC .((((...........((.((((.(((.....)))...)))))).......((((((((.....))))))))........))))................... ( -19.80, z-score = -1.53, R) >consensus UGUUCC_UCCAGCCACGUAGCUCUUCUUGCC_AGACUUGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUACAAAACAGAACCAAUAUCCCCAUUGUCAAC ...................((((.(((.....)))...)))).........((((((((.....))))))))............................... (-16.06 = -15.90 + -0.16)
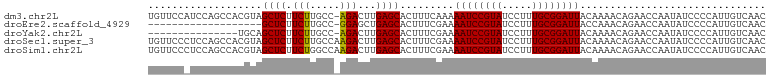
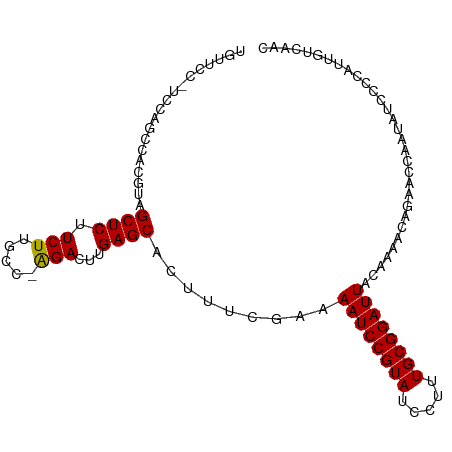
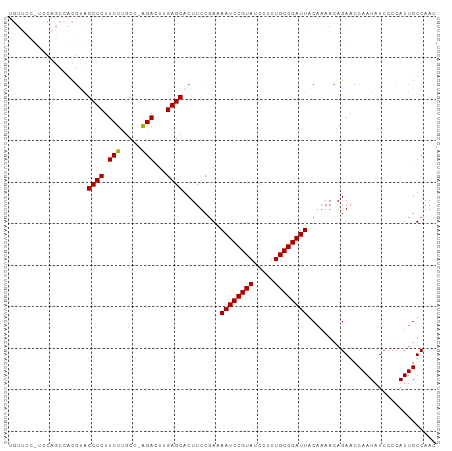
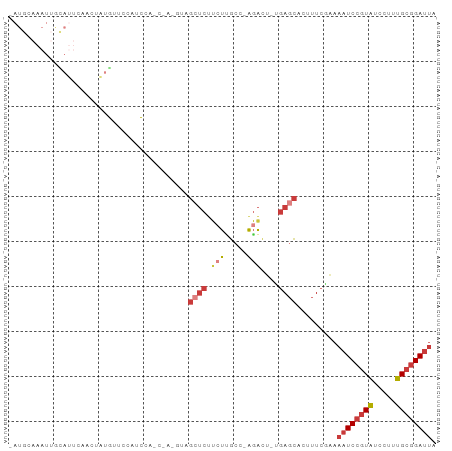
| Location | 14,425,964 – 14,426,056 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 63.90 |
| Shannon entropy | 0.68874 |
| G+C content | 0.42045 |
| Mean single sequence MFE | -19.68 |
| Consensus MFE | -8.86 |
| Energy contribution | -9.90 |
| Covariance contribution | 1.04 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.45 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.48 |
| SVM RNA-class probability | 0.991497 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14425964 92 - 23011544 -AUGCAAAUUGCAUUCAAGUAUGUUCCAUCCAGCCACGUAGCUCUUCUUGCC-AGACU-UGAGCACUUUCAAAAAUCCGUAUCCUUUGCGGAUUA -.((.(((.(((..((((((.((........(((......)))........)-).)))-)))))).)))))..((((((((.....)))))))). ( -20.69, z-score = -2.12, R) >droWil1.scaffold_180764 2698624 84 - 3949147 AAUGCAAAUUAUAACGAUUGUUAUUGAAUU--------CGCUUCAACUGUCCGAUAGUCGGAUAA---UCCGAAAUCCUUUUUCUUAGUCGAAUG ..............((((((...(((((..--------...))))).((((((.....)))))).---...((((.....)))).)))))).... ( -14.60, z-score = -0.86, R) >droAna3.scaffold_12916 14786234 84 + 16180835 -AUGCAAAUGAUAUUAAGCUAUGGACCCUC--------UGGAACUUAUCGGAAAGGCUGAAAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAU-- -.((((((.(((((((((...(((.....)--------))...))))((((((..((.....))..))))))......))))).))))))...-- ( -18.70, z-score = -0.89, R) >droEre2.scaffold_4929 6179314 69 - 26641161 ----------------AUGCAAAUUGCAUU--------CAGCUCUUCUUGCC-GGAGC-UGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUA ----------------.(((.....)))((--------(((((((.......-)))))-))))..........((((((((.....)))))))). ( -23.40, z-score = -3.32, R) >droYak2.chr2L 14530632 77 - 22324452 ----------------AUGCAAAUUGCAUUCAACUAUGCAGCUCUUCUUGCC-AGACU-UGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUA ----------------........(((((......)))))((((.(((....-)))..-.)))).........((((((((.....)))))))). ( -20.30, z-score = -2.70, R) >droSec1.super_3 685393 93 - 7220098 -AUGCAAAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUUGCCAAGACU-UGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUA -.(((.....)))(((...(((((...........)))))((((.(((.....)))..-.))))......)))((((((((.....)))))))). ( -19.50, z-score = -1.80, R) >droSim1.chr2L 14174149 93 - 22036055 -AUGCAAAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUGGCCAAGACU-UGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUA -.(((.....)))(((...(((((...........)))))((((.(((.....)))..-.))))......)))((((((((.....)))))))). ( -20.60, z-score = -1.53, R) >consensus _AUGCAAAUUGCAUUCAACUAUGUUCCAUCCA_C_A_GUAGCUCUUCUUGCC_AGACU_UGAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUUA ........................................((((................)))).........((((((((.....)))))))). ( -8.86 = -9.90 + 1.04)
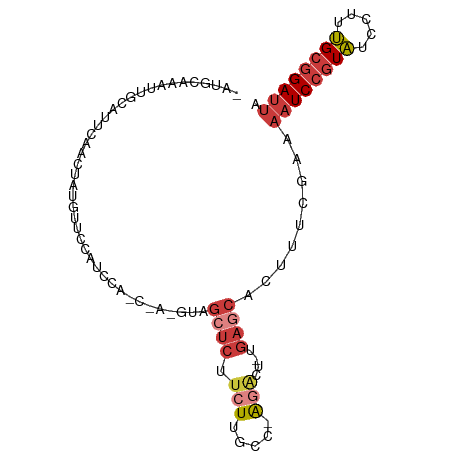
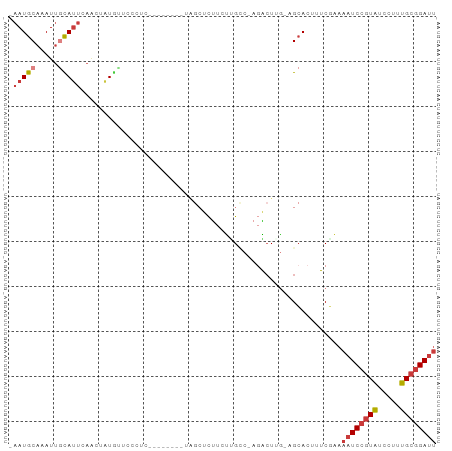
| Location | 14,425,965 – 14,426,057 |
|---|---|
| Length | 92 |
| Sequences | 7 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 67.77 |
| Shannon entropy | 0.61448 |
| G+C content | 0.41805 |
| Mean single sequence MFE | -19.26 |
| Consensus MFE | -7.76 |
| Energy contribution | -8.46 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.03 |
| SVM RNA-class probability | 0.876927 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
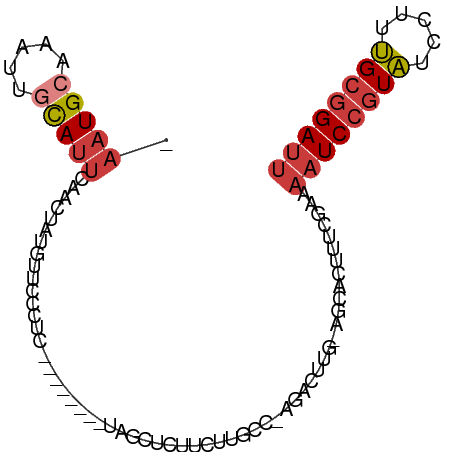
>dm3.chr2L 14425965 92 - 23011544 -AAUGCAAAUUGCAUUCAAGUAUGUUCCAUCCAGCCACGUAGCUCUUCUUGCC-AGACUUG-AGCACUUUCAAAAAUCCGUAUCCUUUGCGGAUU -..((.(((.(((..((((((.((........(((......)))........)-).)))))-)))).)))))..((((((((.....)))))))) ( -19.99, z-score = -1.84, R) >droWil1.scaffold_180764 2698625 84 - 3949147 AAAUGCAAAUUAUAACGAUUGUUAUUGAAU---------UCGCUUCAACUGUCC--GAUAGUCGGAUAAUCCGAAAUCCUUUUUCUUAGUCGAAU ...............((((((...(((((.---------....))))).(((((--(.....))))))....((((.....)))).))))))... ( -14.60, z-score = -0.91, R) >droAna3.scaffold_12916 14786234 85 + 16180835 -AAUGCAAAUGAUAUUAAGCUAUGGACCCUC--------UGGAACUUAUCGGAAAGGCUGAAAGCACUUUCGAAAAUCCGUAUCCUUUGCGGAU- -..((((((.(((((((((...(((.....)--------))...))))((((((..((.....))..))))))......))))).))))))...- ( -18.70, z-score = -0.89, R) >droEre2.scaffold_4929 6179315 69 - 26641161 -AAUGCAAAUUGCAUUCAGCUCU--------------------UCUUGCCGGA---GCUGA--GCACUUUCGAAAAUCCGUAUCCUUUGCGGAUU -.((((.....))))((((((((--------------------.......)))---)))))--...........((((((((.....)))))))) ( -22.90, z-score = -3.05, R) >droYak2.chr2L 14530633 77 - 22324452 -AAUGCAAAUUGCAUUCAACUAUGC---------------AGCUCUUCUUGCC-AGACUUG-AGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUU -.........(((((......))))---------------)((((.(((....-)))...)-))).........((((((((.....)))))))) ( -19.60, z-score = -2.36, R) >droSec1.super_3 685394 93 - 7220098 -AAUGCAAAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUUGCCAAGACUUG-AGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUU -..(((.....)))(((...(((((...........)))))((((.(((.....)))...)-)))......)))((((((((.....)))))))) ( -18.80, z-score = -1.51, R) >droSim1.chr2L 14174150 93 - 22036055 -AAUGCAAAUUGCAUUCAACUAUGUUCCCUCCAGCCACGUAGCUCUUCUGGCCAAGACUUG-AGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUU -(((((.....)))))......(((((..((..((((.(........)))))...))...)-))))........((((((((.....)))))))) ( -20.20, z-score = -1.37, R) >consensus _AAUGCAAAUUGCAUUCAACUAUGUUCCCUC________UAGCUCUUCUUGCC_AGACUUG_AGCACUUUCGAAAAUCCGUAUCCUUUGCGGAUU .(((((.....)))))..........................................................((((((((.....)))))))) ( -7.76 = -8.46 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:11 2011