| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,420,737 – 14,420,827 |
| Length | 90 |
| Max. P | 0.976906 |

| Location | 14,420,737 – 14,420,827 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 94 |
| Reading direction | reverse |
| Mean pairwise identity | 87.58 |
| Shannon entropy | 0.23798 |
| G+C content | 0.54361 |
| Mean single sequence MFE | -29.74 |
| Consensus MFE | -25.57 |
| Energy contribution | -26.73 |
| Covariance contribution | 1.17 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.27 |
| Structure conservation index | 0.86 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.976906 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14420737 90 - 23011544 GAGAACCC-GCAACGCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUA-UUCCAU--UCCACGGCG .......(-((...(((((..((((((((((((((((((....))))))))).))).))))))))))).((.....-......--...)).))) ( -32.54, z-score = -2.72, R) >droSim1.chr2L 14169045 90 - 22036055 GAGAACCC-GCAACGCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUA-UUCCAU--UCCACGGCG .......(-((...(((((..((((((((((((((((((....))))))))).))).))))))))))).((.....-......--...)).))) ( -32.54, z-score = -2.72, R) >droSec1.super_3 680256 90 - 7220098 GAGAACCC-GCAACUCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUA-UUCCAU--UCCACGGCG .......(-((.(((((((..((((((((((((((((((....))))))))).))).)))))))))).))).....-......--......))) ( -29.87, z-score = -2.16, R) >droYak2.chr2L 14525328 91 - 22324452 GAGAACCC-GCAACGCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUACUUCCAU--UCCACUGCG .......(-(((..(((((..((((((((((((((((((....))))))))).))).)))))))))))(((....))).....--.....)))) ( -34.60, z-score = -3.78, R) >droEre2.scaffold_4929 6174229 94 - 26641161 GAGAACCCUGCAACGCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUACUUCCAUCCUCCACUGCC (((...........(((((..((((((((((((((((((....))))))))).))).)))))))))))(((....)))......)))....... ( -32.70, z-score = -3.01, R) >droAna3.scaffold_12916 14781490 77 + 16180835 GAGAACCC-GCAACGCCCCAAACUGUCAGUUAUACAUCACGAUUGAUGGAUGUGCUC--CGGUGCUCCGAUGCCUCGGUG-------------- ........-....((((.....(((((((((.........)))))))))....((..--(((....)))..))...))))-------------- ( -16.20, z-score = 0.79, R) >consensus GAGAACCC_GCAACGCCCCAAACUGUCAGUUAUCCAUCACGAUUGAUGGAUGUGCUCGACGGUGGGGCAGUCUUUA_UUCCAU__UCCACGGCG ..............(((((..((((((((((((((((((....))))))))).))).))))))))))).......................... (-25.57 = -26.73 + 1.17)


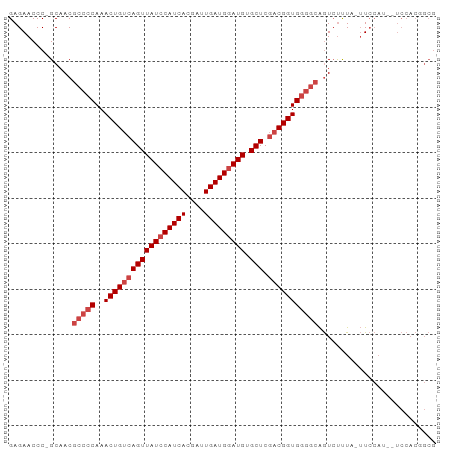
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:07 2011