
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,413,220 – 14,413,355 |
| Length | 135 |
| Max. P | 0.817093 |

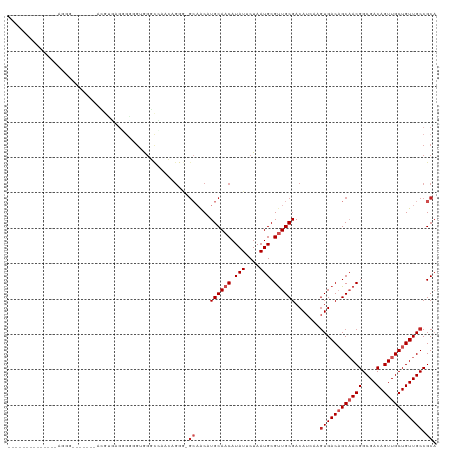
| Location | 14,413,220 – 14,413,333 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | forward |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.59174 |
| G+C content | 0.56061 |
| Mean single sequence MFE | -32.10 |
| Consensus MFE | -19.17 |
| Energy contribution | -19.54 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.80 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.817093 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
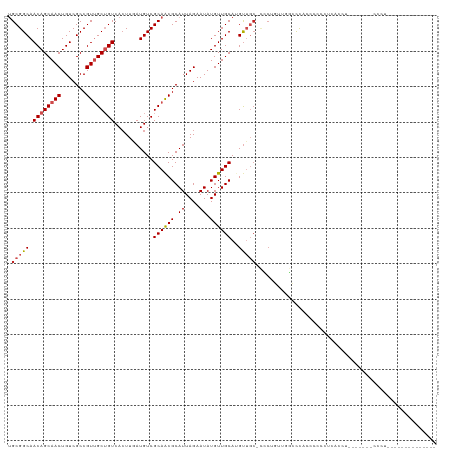
>dm3.chr2L 14413220 113 + 23011544 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGC-CCCUGUUGGCCCACUACGCGCUCGU-------CCCUGCCCAGCAACUGGC .((((((((((((((..((..((((((.(((....)))...)))))).....))....))))).))))))-)..((((((.(....(((....)))-------....).))))))....)) ( -35.10, z-score = -0.07, R) >droSim1.chr2L 14160897 113 + 22036055 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGC-CCCUGUUGGCCCACUACCCGCUCGU-------CCCUGCCCAGCAACUGGC .((((((((((((((..((..((((((.(((....)))...)))))).....))....))))).))))))-)..((((((..........((....-------....))))))))....)) ( -33.90, z-score = -0.35, R) >droSec1.super_3 672723 113 + 7220098 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGC-CCCUGUUGGCCCACUAGCCGCUCGU-------CCCUGCCCAGCAACUGGC .((((((((((((((..((..((((((.(((....)))...)))))).....))....))))).))))))-)......(((......)))))....-------......((((...)))). ( -36.00, z-score = -0.47, R) >droYak2.chr2L 14517731 113 + 22324452 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGC-CC-UGCUGGCCCACUCCCCUCACGUU------CCCUGCCCUUCAACUGGC .((((((((((((((..((..((((((.(((....)))...)))))).....))....))))).))))))-).-.))....................------....(((........))) ( -30.10, z-score = -0.23, R) >droEre2.scaffold_4929 6166536 120 + 26641161 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGCUGC-CCCUGUUGGCCCACACCCCCCACCGCCCCCCUCCCUUCCUUGCAACUGGC (((((((((((((((.......)))))))).......)))))))..............((((((......-.......(((..............))).............)))))).... ( -28.32, z-score = 0.20, R) >droAna3.scaffold_12916 14774537 106 - 16180835 UGCGGCAACAGCAACUGUCGCCGUGGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGCCUUC-CCACAGCCGCCUCACUCGUCUCUCGCACGGGC-------------- .(((((.((((...)))).)))))((((((.....((....((((((((.((.....)).)))..)))))..))-..))))))((((....((.....))...))))-------------- ( -31.50, z-score = 0.21, R) >droPer1.super_5 1725370 108 + 6813705 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAGCGCAUUAGAAUAUGUUGCAUGUUGCUGUCGCUUCGGCUUCCCCUCUGGCCGCUGCUGUUGCUG------------- .((((((((((((((..((.....((((((.((.....)).)))))).....))....))))).))))))))).((..(((((........)))))..))........------------- ( -41.20, z-score = -1.89, R) >dp4.chr4_group1 1781448 102 - 5278887 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAGCGCAUUAGAAUAUGUUGCAUGUUGCUGUCGCUUCGGCUUCCCCUCUGGCCGCUGCUG------------------- .((((((((((((((..((.....((((((.((.....)).)))))).....))....))))).))))))))).((..(((((........)))))..))..------------------- ( -41.20, z-score = -2.65, R) >droWil1.scaffold_180764 2671896 90 + 3949147 UGCGGCAACAGCAACUGUCGCCGUUGCUGUCAGUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAGGUU-CCCUCGCUCUCGUUUCGUUGUC------------------------------ (((((((((((((((.......)))))))).......)))))))..(((...(((.(((..(((((..-.))).))...))))))..))).------------------------------ ( -26.21, z-score = -1.28, R) >droVir3.scaffold_12723 3024221 90 - 5802038 AGCGGCAACAGCAACUGUCGCCGUUGCAGUCACUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAAGUG---CCCCGUCUGACCCCCCCAUAAA---------------------------- .(.((((...(((((..((..((((((.(.((.....)).))))))).....))....)))))...))---)).)..................---------------------------- ( -22.00, z-score = -1.03, R) >droMoj3.scaffold_6500 16319692 98 - 32352404 AGCGGCAACAGCAACUGUCGCCGUGGCAGUCACUUCAUGUCGCAACGCAUUAGAAUAUGUUGCAAGUAGCCCCCCGUCUGACCUGCACACAUAUUAAA----------------------- .(((((.((((...)))).)))((((((.........))))))...)).....(((((((((((.((.((.....))...)).)))).)))))))...----------------------- ( -27.60, z-score = -1.23, R) >consensus UGCGGCAACAGCAACUGUCGCCGUUGCUGUCACUUGAUGUCGCAACGCAUUAGAAUAUGUUGCAUGUUGC_CCCUGUUGGCCCACCCCCCUCACCU_______CCCU______________ .((((((((((((((.......)))))))).......))))))...(((.((.....)).))).......................................................... (-19.17 = -19.54 + 0.36)

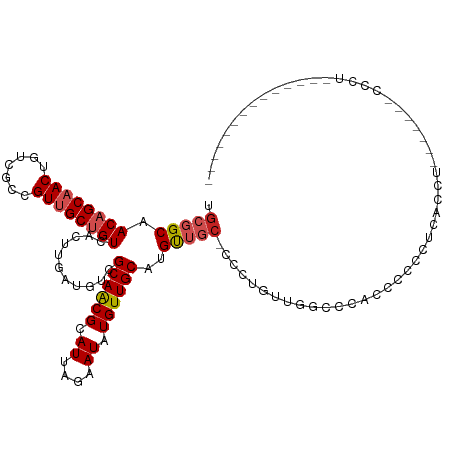
| Location | 14,413,220 – 14,413,333 |
|---|---|
| Length | 113 |
| Sequences | 11 |
| Columns | 121 |
| Reading direction | reverse |
| Mean pairwise identity | 71.31 |
| Shannon entropy | 0.59174 |
| G+C content | 0.56061 |
| Mean single sequence MFE | -36.61 |
| Consensus MFE | -20.69 |
| Energy contribution | -21.60 |
| Covariance contribution | 0.91 |
| Combinations/Pair | 1.00 |
| Mean z-score | -0.79 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.739908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
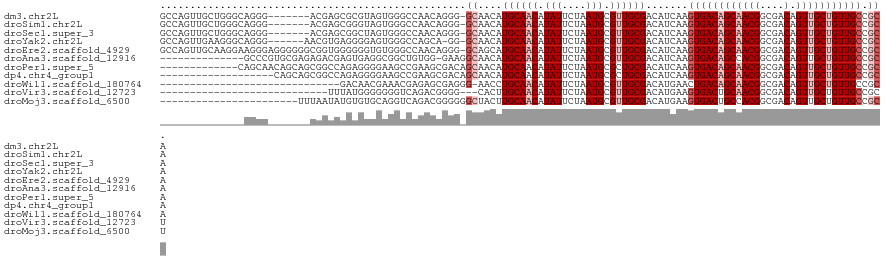
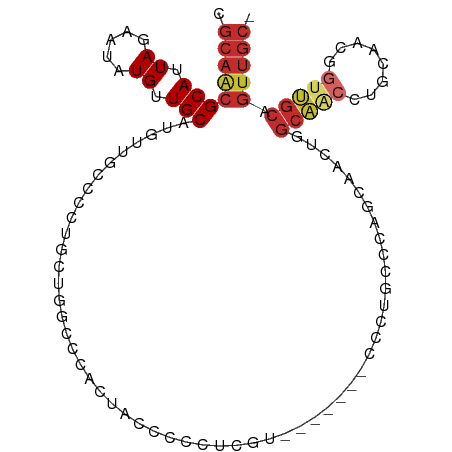
>dm3.chr2L 14413220 113 - 23011544 GCCAGUUGCUGGGCAGGG-------ACGAGCGCGUAGUGGGCCAACAGGG-GCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ((..((((((((((....-------(((....))).....)))....(..-(((((..(((...........))))))))..).........)))))))((((((((...)))))))))). ( -40.70, z-score = -0.25, R) >droSim1.chr2L 14160897 113 - 22036055 GCCAGUUGCUGGGCAGGG-------ACGAGCGGGUAGUGGGCCAACAGGG-GCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA (((..(((((.(.(....-------....)).)))))..))).....(.(-((((((.(((((.........(((((..(........)..)).)))..(....).)))))))))))).). ( -39.30, z-score = -0.13, R) >droSec1.super_3 672723 113 - 7220098 GCCAGUUGCUGGGCAGGG-------ACGAGCGGCUAGUGGGCCAACAGGG-GCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ((((((((((.(......-------.).)))))))....))).....(.(-((((((.(((((.........(((((..(........)..)).)))..(....).)))))))))))).). ( -42.40, z-score = -0.86, R) >droYak2.chr2L 14517731 113 - 22324452 GCCAGUUGAAGGGCAGGG------AACGUGAGGGGAGUGGGCCAGCA-GG-GCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ((..((((.((.(((.((------((..((.((........)).(((-.(-....).)))..))..))))..))).)).)))).....(..(((((((((....).))))))))..).)). ( -39.00, z-score = -0.74, R) >droEre2.scaffold_4929 6166536 120 - 26641161 GCCAGUUGCAAGGAAGGGAGGGGGGCGGUGGGGGGUGUGGGCCAACAGGG-GCAGCAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ((..(((((((.(.......((((...(((..(.((((..(((......)-)).)))).)..))).))))....).))))))).....(..(((((((((....).))))))))..).)). ( -45.80, z-score = -1.65, R) >droAna3.scaffold_12916 14774537 106 + 16180835 --------------GCCCGUGCGAGAGACGAGUGAGGCGGCUGUGG-GAAGGCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCCACGGCGACAGUUGCUGUUGCCGCA --------------(((((..((.....))..)).)))((((((.(-...(....).((((((.(((....))).))))))........).)))))).(((((((((...))))))))).. ( -39.40, z-score = -0.90, R) >droPer1.super_5 1725370 108 - 6813705 -------------CAGCAACAGCAGCGGCCAGAGGGGAAGCCGAAGCGACAGCAACAUGCAACAUAUUCUAAUGCGCUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA -------------..(((((((((((..((....))...((((..((....)).....(((...........)))((((..((.....))..))))..))))....))))))))))).... ( -37.20, z-score = -1.23, R) >dp4.chr4_group1 1781448 102 + 5278887 -------------------CAGCAGCGGCCAGAGGGGAAGCCGAAGCGACAGCAACAUGCAACAUAUUCUAAUGCGCUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA -------------------..((.(((((((...((((.((....))....((.....))......))))..)).)))))........(..(((((((((....).))))))))..).)). ( -34.90, z-score = -0.87, R) >droWil1.scaffold_180764 2671896 90 - 3949147 ------------------------------GACAACGAAACGAGAGCGAGGG-AACCUGCAACAUAUUCUAAUGCGUUGCGACAUGAACUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ------------------------------...............(((.(..-..).)))............((((((((..((.....))...)))))((((((((...))))))))))) ( -24.90, z-score = -0.94, R) >droVir3.scaffold_12723 3024221 90 + 5802038 ----------------------------UUUAUGGGGGGGUCAGACGGGG---CACUUGCAACAUAUUCUAAUGCGUUGCGACAUGAAGUGACUGCAACGGCGACAGUUGCUGUUGCCGCU ----------------------------((((((.....(((......))---)..(((((((.(((....))).)))))))))))))(((...((((((((((...))))))))))))). ( -26.80, z-score = -0.30, R) >droMoj3.scaffold_6500 16319692 98 + 32352404 -----------------------UUUAAUAUGUGUGCAGGUCAGACGGGGGGCUACUUGCAACAUAUUCUAAUGCGUUGCGACAUGAAGUGACUGCCACGGCGACAGUUGCUGUUGCCGCU -----------------------...(((((((.(((((((.((........)))))))))))))))).....((((..(........)..)).))..(((((((((...))))))))).. ( -32.30, z-score = -0.77, R) >consensus ______________AGGG_______ACGUGAGGGGGGUGGGCCAACAGGG_GCAACAUGCAACAUAUUCUAAUGCGUUGCGACAUCAAGUGACAGCAACGGCGACAGUUGCUGUUGCCGCA ...................................................((....((((((.(((....))).)))))).......((((((((((((....).))))))))))).)). (-20.69 = -21.60 + 0.91)
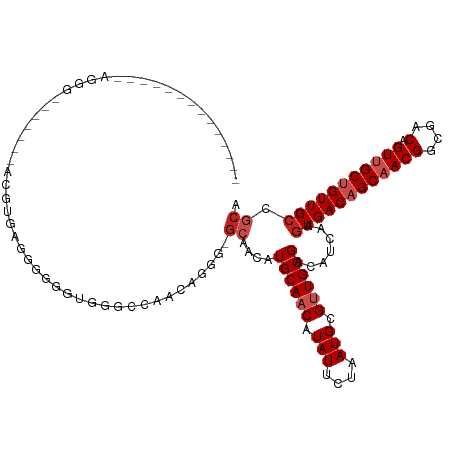
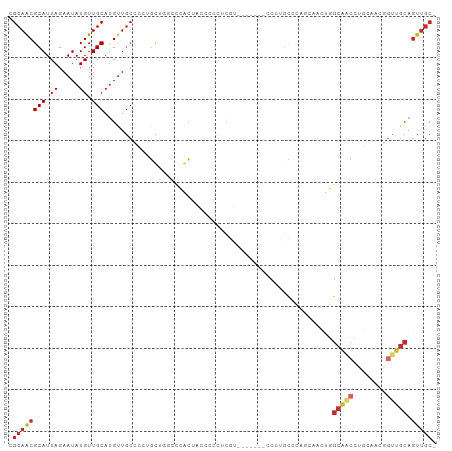
| Location | 14,413,260 – 14,413,355 |
|---|---|
| Length | 95 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 65.95 |
| Shannon entropy | 0.69658 |
| G+C content | 0.56772 |
| Mean single sequence MFE | -29.07 |
| Consensus MFE | -11.68 |
| Energy contribution | -12.44 |
| Covariance contribution | 0.77 |
| Combinations/Pair | 1.20 |
| Mean z-score | -0.97 |
| Structure conservation index | 0.40 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.586122 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14413260 95 + 23011544 CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCCCUGUUGGCCCACUACGCGCUCGU-------CCCUGCCCAGCAACUGGCAACCUGCAACGGUUGCAGUUGC- .((((((((........(((((((.((((((..((((((.(....(((....)))-------....).))))))...)))))).)))))))..))).)))))- ( -34.50, z-score = -1.86, R) >droSim1.chr2L 14160937 95 + 22036055 CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCCCUGUUGGCCCACUACCCGCUCGU-------CCCUGCCCAGCAACUGGCAACCUGCAACGGUUGCAGUUGC- .((((((((........(((((((.((((((..((((((..........((....-------....))))))))...)))))).)))))))..))).)))))- ( -33.30, z-score = -2.12, R) >droSec1.super_3 672763 95 + 7220098 CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCCCUGUUGGCCCACUAGCCGCUCGU-------CCCUGCCCAGCAACUGGCAACCUGCAACGGUUGCAGUUGC- .((((((((........(((((((.((((((..(((((((......)))((....-------....))..))))...)))))).)))))))..))).)))))- ( -33.80, z-score = -1.73, R) >droYak2.chr2L 14517771 95 + 22324452 CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCC-UGCUGGCCCACUCCCCUCACGUU------CCCUGCCCUUCAACUGGCAACCUGCAACGGUUGCAGUUGC- .((((((((........(((((((.((((((.-((..(((................------....)))...))...)))))).)))))))..))).)))))- ( -27.45, z-score = -1.36, R) >droEre2.scaffold_4929 6166576 102 + 26641161 CGCAACGCAUUAGAAUAUGUUGCAUGCUGCCCCUGUUGGCCCACACCCCCCACCGCCCCCCUCCCUUCCUUGCAACUGGCAACCUGCAACGGUUGCAGUUGC- .((((((.((....)).)))))).....(((......)))...............................(((((((.(((((......))))))))))))- ( -25.90, z-score = -0.81, R) >droAna3.scaffold_12916 14774577 82 - 16180835 CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCU------UCCCACAGCCGCCUCAC-------UCGUCUCUCGCA-CGGGCA--CUG----GGUUGCAGUUGC- .((((((((.((.....)).)))....(((..------.((((.....((((...-------.((.....))..-.)))).--.))----))..))))))))- ( -20.90, z-score = 0.77, R) >droPer1.super_5 1725410 92 + 6813705 CGCAGCGCAUUAGAAUAUGUUGCAUGUUGCUGUCGCU--UCGGCUUCCCCUCUGG--------C-CGCUGCUGUUGCUGCUGCACCAAUGGGUGGCAGUUGCA .(((((((..(((((((((...)))))).)))..)).--..((((........))--------)-)))))).((.(((((..(.((...)))..))))).)). ( -36.10, z-score = -0.71, R) >dp4.chr4_group1 1781488 86 - 5278887 CGCAGCGCAUUAGAAUAUGUUGCAUGUUGCUGUCGCU--UCGGCUUCCCCUCUGG--------C-CGC------UGCUGCUGCAGCAAUGGGUGGCAGUUGCA .((((((.((....)).)))))).(((.(((((((((--(.((((........))--------)-)((------(((....)))))...)))))))))).))) ( -36.60, z-score = -1.15, R) >droWil1.scaffold_180764 2671936 73 + 3949147 CGCAACGCAUUAGAAUAUGUUGCAGGUUCCC-UCGCU--CUCGUUUCGUUGUCAU--------CUCGAAAAGGUUGCAGUUUCA------------------- .(((((((((......)))).(((((...))-).)).--....(((((.......--------..)))))..))))).......------------------- ( -13.10, z-score = 0.23, R) >consensus CGCAACGCAUUAGAAUAUGUUGCAUGUUGCCCCUGCUGGCCCACUACCCCCUCGU_______CCCUGCCCAGCAACUGGCAACCUGCAACGGUUGCAGUUGC_ .((((((((.((.....)).))).......................................................(((((........))))).))))). (-11.68 = -12.44 + 0.77)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:06 2011