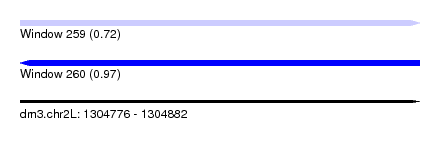
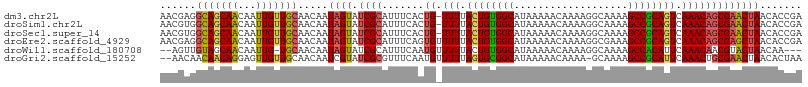
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,304,776 – 1,304,882 |
| Length | 106 |
| Max. P | 0.969934 |

| Location | 1,304,776 – 1,304,882 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | forward |
| Mean pairwise identity | 87.81 |
| Shannon entropy | 0.23362 |
| G+C content | 0.39941 |
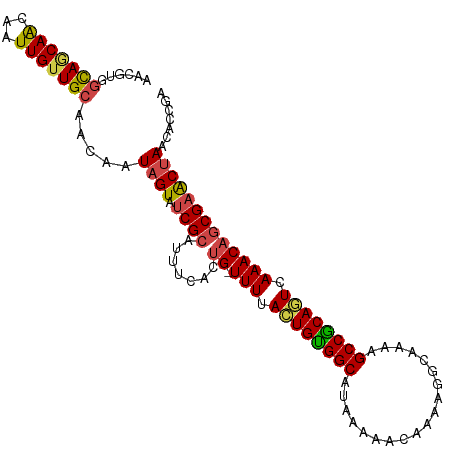
| Mean single sequence MFE | -30.40 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.72 |
| Covariance contribution | 0.61 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.66 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.49 |
| SVM RNA-class probability | 0.718091 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1304776 106 + 23011544 UCGGUGUUAGUUCGCUGUUUGACUGCGGCUUUUGCCUUUUGUUUUUAUGCCACAGUAAAA-CAGUGAAAUGCGAUACUAUUGUUGCAACAAUUGUUGCUGCCUCGUU ..((((((..((((((((((.((((.(((....((.....))......))).)))).)))-)))))))....))))))...((.(((((....))))).))...... ( -34.30, z-score = -3.30, R) >droSim1.chr2L 1276313 106 + 22036055 UCGGUGUUAGUUCGCUGUUUGACUGCGGCUUUUGCCUUUUGUUUUUAUGCCACAGUAAAA-CAGUGAAAUGCGAUACUAUUGUUGCAACAAUUGUUGCUGCCACGUU ..((((((..((((((((((.((((.(((....((.....))......))).)))).)))-)))))))....))))))...((.(((((....))))).))...... ( -34.30, z-score = -2.91, R) >droSec1.super_14 1254547 106 + 2068291 UCGGUGUUAGUUCGCUGUUUGACUGCGGCUUUUGCCUUUUGUUUUUAUGCCACAGUAAAA-CAGUGAAAUGCGAUACUAUUGUUGCAACAAUUGUUGCUGCCACGUU ..((((((..((((((((((.((((.(((....((.....))......))).)))).)))-)))))))....))))))...((.(((((....))))).))...... ( -34.30, z-score = -2.91, R) >droEre2.scaffold_4929 1343582 107 + 26641161 UCGGUGUUAGCUCGCUGUUUGACUGCGGCUUUCGCCUUUUGUUUUUAUGCCACAGUAAAAACACUGAAAUGCGAUACUAUUGUUGCAACAAUUGUUGCUGCCUCGUU ..((((..(((.(((.(.....).))))))..))))...(((((((((......))))))))).......((((.......((.(((((....))))).)).)))). ( -28.60, z-score = -1.46, R) >droWil1.scaffold_180708 9629323 101 - 12563649 ---UUGUUAGUACGUUGUUUGAAUGUGGCUUUUGCCUUUUGUUUUUAUGCCACAGUAAAAACAUUGAAAUGCGAUACUAUUGUUGCA-CAAUUGUUGCUACAACU-- ---....((((((((..((..(....(((....)))...(((((((((......))))))))))..))..))).)))))((((.(((-(....).))).))))..-- ( -22.90, z-score = -1.01, R) >droGri2.scaffold_15252 6810330 104 - 17193109 UUAGUGUUAGUUCGCAGUUUGAAUGCGGCUUUUGC-UUUUGUUUUUAUGCCGCACUAAAAACAUUGAAACGCGAUACGAUUGUUGCAACAACUCCUGUUGUUGUU-- ...(((((.(((((..((((...((((((....((-....))......))))))....))))..)).)))..))))).......((((((((....)))))))).-- ( -28.00, z-score = -1.59, R) >consensus UCGGUGUUAGUUCGCUGUUUGACUGCGGCUUUUGCCUUUUGUUUUUAUGCCACAGUAAAA_CAGUGAAAUGCGAUACUAUUGUUGCAACAAUUGUUGCUGCCACGUU ..(((..((((((((.((((.((((.(((....)))......((((((......)))))).)))).)))))))).)))).....(((((....))))).)))..... (-20.10 = -20.72 + 0.61)
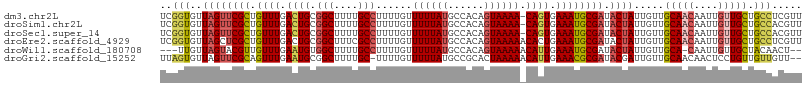
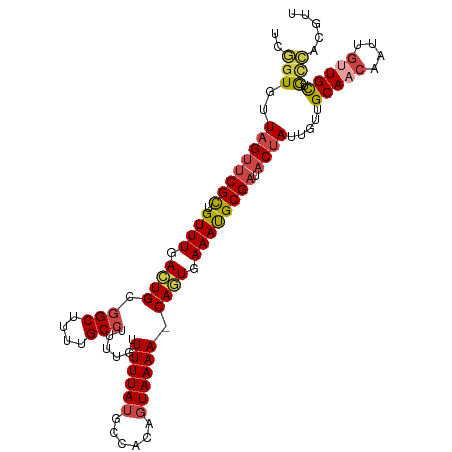
| Location | 1,304,776 – 1,304,882 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 87.81 |
| Shannon entropy | 0.23362 |
| G+C content | 0.39941 |
| Mean single sequence MFE | -28.40 |
| Consensus MFE | -20.61 |
| Energy contribution | -20.75 |
| Covariance contribution | 0.14 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.82 |
| SVM RNA-class probability | 0.969934 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1304776 106 - 23011544 AACGAGGCAGCAACAAUUGUUGCAACAAUAGUAUCGCAUUUCACUG-UUUUACUGUGGCAUAAAAACAAAAGGCAAAAGCCGCAGUCAAACAGCGAACUAACACCGA .....((..(((((....))))).....((((.((((.......((-(((.((((((((...................)))))))).)))))))))))))...)).. ( -30.32, z-score = -3.32, R) >droSim1.chr2L 1276313 106 - 22036055 AACGUGGCAGCAACAAUUGUUGCAACAAUAGUAUCGCAUUUCACUG-UUUUACUGUGGCAUAAAAACAAAAGGCAAAAGCCGCAGUCAAACAGCGAACUAACACCGA ...(((((.(((((....))))).((....))...))..(((.(((-(((.((((((((...................)))))))).)))))).)))....)))... ( -32.11, z-score = -3.45, R) >droSec1.super_14 1254547 106 - 2068291 AACGUGGCAGCAACAAUUGUUGCAACAAUAGUAUCGCAUUUCACUG-UUUUACUGUGGCAUAAAAACAAAAGGCAAAAGCCGCAGUCAAACAGCGAACUAACACCGA ...(((((.(((((....))))).((....))...))..(((.(((-(((.((((((((...................)))))))).)))))).)))....)))... ( -32.11, z-score = -3.45, R) >droEre2.scaffold_4929 1343582 107 - 26641161 AACGAGGCAGCAACAAUUGUUGCAACAAUAGUAUCGCAUUUCAGUGUUUUUACUGUGGCAUAAAAACAAAAGGCGAAAGCCGCAGUCAAACAGCGAGCUAACACCGA .....((..(((((....))))).....((((.((((.......((((((((........))))))))...(((....)))...........))))))))...)).. ( -29.40, z-score = -2.07, R) >droWil1.scaffold_180708 9629323 101 + 12563649 --AGUUGUAGCAACAAUUG-UGCAACAAUAGUAUCGCAUUUCAAUGUUUUUACUGUGGCAUAAAAACAAAAGGCAAAAGCCACAUUCAAACAACGUACUAACAA--- --.(((((........(((-(((....((((((..((((....))))...)))))).))))))........(((....)))........)))))..........--- ( -22.70, z-score = -2.25, R) >droGri2.scaffold_15252 6810330 104 + 17193109 --AACAACAACAGGAGUUGUUGCAACAAUCGUAUCGCGUUUCAAUGUUUUUAGUGCGGCAUAAAAACAAAA-GCAAAAGCCGCAUUCAAACUGCGAACUAACACUAA --..(((((((....)))))))........((.(((((.......((((..((((((((............-......)))))))).)))))))))))......... ( -23.78, z-score = -1.57, R) >consensus AACGUGGCAGCAACAAUUGUUGCAACAAUAGUAUCGCAUUUCACUG_UUUUACUGUGGCAUAAAAACAAAAGGCAAAAGCCGCAGUCAAACAGCGAACUAACACCGA ......(((((((...))))))).....((((.((((.......((.(((.((((((((...................)))))))).)))))))))))))....... (-20.61 = -20.75 + 0.14)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:15 2011