| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,400,452 – 14,400,543 |
| Length | 91 |
| Max. P | 0.991738 |

| Location | 14,400,452 – 14,400,543 |
|---|---|
| Length | 91 |
| Sequences | 12 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 76.27 |
| Shannon entropy | 0.50706 |
| G+C content | 0.35819 |
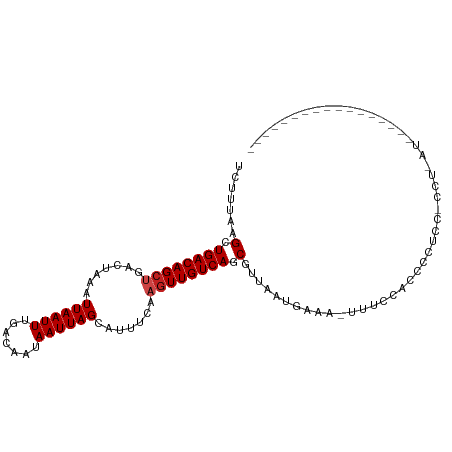
| Mean single sequence MFE | -18.42 |
| Consensus MFE | -10.50 |
| Energy contribution | -10.75 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.26 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.50 |
| SVM RNA-class probability | 0.991738 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 14400452 91 + 23011544 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAA-UUUUCACCCCUCCACCUGAUG---------------- ((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))..-.....................---------------- ( -19.04, z-score = -3.15, R) >droSim1.chr2L 14148240 91 + 22036055 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAA-UUUCCACCCCUCCACCUGAUG---------------- ((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))..-.....................---------------- ( -19.04, z-score = -3.36, R) >droSec1.super_3 660055 90 + 7220098 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAA--UUUCCACCCCUUCACCUGAUG---------------- ((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).)).--.....................---------------- ( -19.04, z-score = -3.12, R) >droYak2.chr2L 14504864 93 + 22324452 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAA-UUUCCACCCCUCCCCGCCCGAUG-------------- ((.((((((((((((((.......((((((.......)))))).......)))))))))).)))).))..-.......................-------------- ( -19.04, z-score = -3.62, R) >droEre2.scaffold_4929 6153615 104 + 26641161 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAAAUUUCCACACCUCCCGCCCUCCCUCCAACUGAUG---- .......((((((((((.......((((((.......)))))).......))))))))))(..(((....)))..)............................---- ( -19.24, z-score = -3.03, R) >droAna3.scaffold_12916 14763211 107 - 16180835 UCUUUAAGCUGACAGCCGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAA-UCGCCCCUCUACCCCCCAAGCAUCUCCUGCCCCGAUG ((.(((((((((((((........((((((.......))))))........))))))))).)))).))..-...................(((.....)))....... ( -17.59, z-score = -2.18, R) >dp4.chr4_group1 1762518 80 - 5278887 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUCCAAGUUGUCAACGUUAAUGAAAUUUCUCGCUG---------------------------- ......(((((((((((.......((((((.......)))))).......))))))))........((......))))).---------------------------- ( -13.04, z-score = -1.00, R) >droPer1.super_12401 209 80 + 712 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAACGUUAAUGAAAUUUCUCGCUG---------------------------- ......(((((((((((.......((((((.......)))))).......))))))))........((......))))).---------------------------- ( -13.04, z-score = -0.76, R) >droWil1.scaffold_180764 2651849 84 + 3949147 UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAACGUUAAUGAAAUUCUGCGGUGAGUG------------------------ .......(((.((.((.((....((((((((((((((..............)))))))).)))))).....)).)).)).))).------------------------ ( -17.54, z-score = -1.20, R) >droVir3.scaffold_12723 3009660 95 - 5802038 UAUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAUUCCGCGGCGAGUGUUGCUCAUUUUG------------- ((((...((((((((((.......((((((.......)))))).......))))))))))...))))......(((((....)))))........------------- ( -23.24, z-score = -1.64, R) >droMoj3.scaffold_6500 16300019 95 - 32352404 AAUUUAAGCUGACAGGCGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGCUAAUGAAUGCGACGGCAACUGUUGCUCCUAUUG------------- ((((((.(((....)))...)))))).......(((((...((((.....(((((((..(((........)))..)))))))..))))..)))))------------- ( -23.20, z-score = -1.60, R) >droGri2.scaffold_15126 705766 76 + 8399593 UAUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCAUUUUUGCCAAUUUUG-------------------------------- .......((((((((((.......((((((.......)))))).......))))))))))................-------------------------------- ( -17.94, z-score = -2.43, R) >consensus UCUUUAAGCUGACAGCUGACUAAAUUAAUUUGACAAUAAUUAGCAUUUCAAGUUGUCAGCGUUAAUGAAA_UUUCCACCCCUCC_CCU_AU_________________ .......(.((((((((.......((((((.......)))))).......)))))))).)................................................ (-10.50 = -10.75 + 0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:40:00 2011