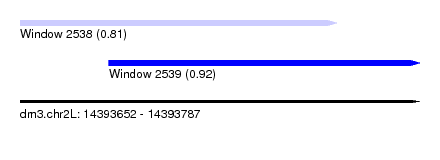
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,393,652 – 14,393,787 |
| Length | 135 |
| Max. P | 0.917403 |

| Location | 14,393,652 – 14,393,759 |
|---|---|
| Length | 107 |
| Sequences | 7 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 78.35 |
| Shannon entropy | 0.42811 |
| G+C content | 0.32678 |
| Mean single sequence MFE | -21.84 |
| Consensus MFE | -12.43 |
| Energy contribution | -12.73 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.41 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.57 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.77 |
| SVM RNA-class probability | 0.811379 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
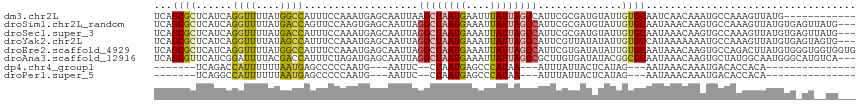
>dm3.chr2L 14393652 107 + 23011544 -AUGUGCACAACUAAUGUUAAU-AACAUUUAAUCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAG---CAAUUAAGCUAAUGAAUUUAUUAGCCAUUCGCGAUGUAU -..((((((....(((((....-.)))))......((((((((..((..............))..)))))---)......(((((((....))))))).....))).))))) ( -20.94, z-score = -0.62, R) >droSim1.chr2L_random 564197 107 + 909653 -AUGUGCACAACUAAUGUUAAU-AACAUUUAAUCAGCGCUCAUCAGGUUUUAUGACCAGUUCCAAGUGAG---CAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUGUAU -..((((......(((((....-.)))))......)))).((((.((((....)))).((((.....)))---).....((((((((....))))))))......))))... ( -26.30, z-score = -2.24, R) >droSec1.super_3 653072 107 + 7220098 -AUGUGCACAACUAAUGUUAAU-AACAUUUAAUCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAG---CAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUGUAU -..((((((....(((((....-.)))))......((((((((..((..............))..)))))---).....((((((((....))))))))....))).))))) ( -25.14, z-score = -2.27, R) >droYak2.chr2L 14497823 107 + 22324452 -AUGUGCACAGCUAAUGUUAAU-AACAUUUAAUCAGCGCUCAUCAGGUUUUAUAGCCAUUUCCAAAUGAG---CAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUUAUAUAU -(((((.((.(((...(((((.-.....))))).)))((((((..((..............))..)))))---).....((((((((....))))))))....))))))).. ( -24.04, z-score = -2.24, R) >droEre2.scaffold_4929 6146822 108 + 26641161 AUUGUGUACAACUAAUGUUAAU-AACAUUUAAUCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAG---CAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUGAUAUAU ..(((....(((....)))...-.)))....((((..((((((..((..............))..)))))---).....((((((((....)))))))).....)))).... ( -22.64, z-score = -1.42, R) >droAna3.scaffold_12916 14757206 107 - 16180835 -GUAACCAGAACUCAUGCUAAU-AGUAUUUAAUCAGCGUUCAUCGGAUUUUACGACCAUUUCUAGAUGAG---CAAUUAGGCUAAUGAAAUUAUUAGCCGCUUGUGAUAUAC -.....................-.((((...((.(((((((((((((.............))).))))))---).....((((((((....))))))))))).))...)))) ( -25.02, z-score = -2.23, R) >dp4.chr4_group1 1753034 92 - 5278887 -CUUUCUAAAAAUAAUGCUUAUUAAUAUUUAAUCAG-----ACC--AUUUUUUAAUGAGCCCCCAAUGAAUUCCUAAUGAGCCCAUAAAUUUAUUACUCA------------ -...............(((((((((...........-----...--.....)))))))))....((((((((....(((....))).)))))))).....------------ ( -8.81, z-score = -1.02, R) >consensus _AUGUGCACAACUAAUGUUAAU_AACAUUUAAUCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAG___CAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUAUAU ...((((......((((((....))))))......))))......((((....))))......................((((((((....))))))))............. (-12.43 = -12.73 + 0.30)
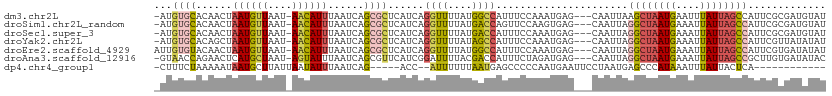
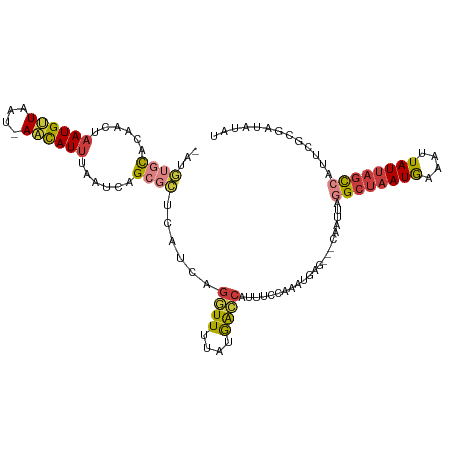
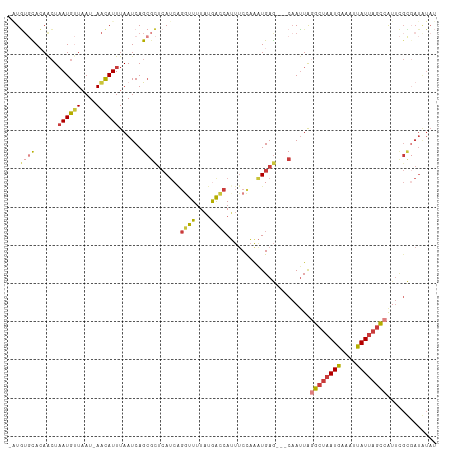
| Location | 14,393,682 – 14,393,787 |
|---|---|
| Length | 105 |
| Sequences | 8 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 67.33 |
| Shannon entropy | 0.62830 |
| G+C content | 0.37006 |
| Mean single sequence MFE | -22.38 |
| Consensus MFE | -11.34 |
| Energy contribution | -11.15 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.44 |
| Mean z-score | -1.22 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.26 |
| SVM RNA-class probability | 0.917403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
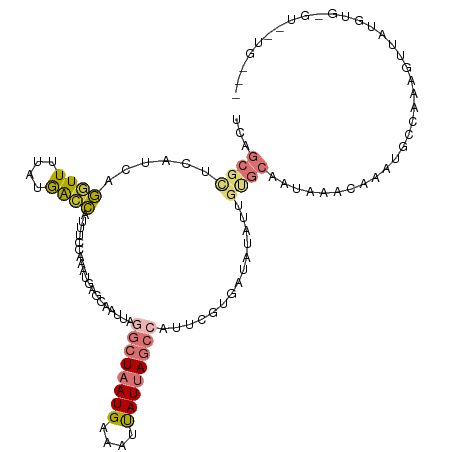
>dm3.chr2L 14393682 105 + 23011544 UCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAAGCUAAUGAAUUUAUUAGCCAUUCGCGAUGUAUUGUGCAAUCAACAAAUGCCAAAGUUAUG------------ ...((((.((((.((((....))))............(((((...(((((((....))))))).))).))))))....)))).......................------------ ( -20.40, z-score = -0.09, R) >droSim1.chr2L_random 564227 114 + 909653 UCAGCGCUCAUCAGGUUUUAUGACCAGUUCCAAGUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUGUAUUGUGCAAUAAACAAGUGCCAAAGUUAUGUGAGUUAUG--- .....((((((..((..((......))..))..)))))).....((((((((....))))))))(((((((.....(((.(((.........)))))).....)))))))....--- ( -27.90, z-score = -1.49, R) >droSec1.super_3 653102 114 + 7220098 UCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGCGAUGUAUUGUGCAAUAAACAAGUGCCAAAGUUAUGUGAGUUAUG--- .....((((((..((..............))..)))))).....((((((((....))))))))(((((((.....(((.(((.........)))))).....)))))))....--- ( -26.74, z-score = -1.33, R) >droYak2.chr2L 14497853 114 + 22324452 UCAGCGCUCAUCAGGUUUUAUAGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUUAUAUAUUGUGCCAUAAAAAAAUGCCAAAGUUAUGUGAGUAGUG--- .....((((((..((..............))..)))))).....((((((((....)))))))).....(((((((...((.(((......))).))....)))))))......--- ( -24.44, z-score = -1.57, R) >droEre2.scaffold_4929 6146853 117 + 26641161 UCAGCGCUCAUCAGGUUUUAUGGCCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUGAUAUAUUGUGCAAUAAACAAGUGCCAGACUUAUGUGGGUGGUGGUG (((.(((((((.(((((...(((((...................((((((((....))))))))............((((.......))))).)))))))))..))))))).))).. ( -30.60, z-score = -0.70, R) >droAna3.scaffold_12916 14757236 114 - 16180835 UCAGCGUUCAUCGGAUUUUACGACCAUUUCUAGAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCGCUUGUGAUAUACGGCGCAAUAAACAAGUGCUAUGGCAAUGGGCAUGUCA--- .....((((((((((.............))).))))))).....((((((((....))))))))(((..(....((..((((.........))))..))...)..)))......--- ( -31.62, z-score = -1.51, R) >dp4.chr4_group1 1753065 84 - 5278887 -------UCAGACCAUUUUUUAAUGAGCCCCCAAUG---AAUUC--CUAAUGAGCCCAUAA---AUUUAUUACUCAUAG---AAUAAACAAAUGACACCACA--------------- -------(((........(((.(((((.....((((---((((.--...(((....))).)---))))))).))))).)---))........))).......--------------- ( -6.99, z-score = -1.38, R) >droPer1.super_5 1697154 84 + 6813705 -------UCAGGCCAUUUUUUAAUGAGCCCCCAAUG---AAUUC--CUAAUGAGCCCAUAA---AUUUAUUACUCAUAG---AAUAAACAAAUGACACCACA--------------- -------((((((((((....)))).))).......---.((((--...(((((.......---........))))).)---))).......))).......--------------- ( -10.36, z-score = -1.69, R) >consensus UCAGCGCUCAUCAGGUUUUAUGACCAUUUCCAAAUGAGCAAUUAGGCUAAUGAAAUUAUUAGCCAUUCGUGAUAUAUUGUGCAAUAAACAAAUGCCAAAGUUAUGUG_GU__UG___ ...((((......((((....))))...................((((((((....))))))))..............))))................................... (-11.34 = -11.15 + -0.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:56 2011