
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,388,800 – 14,388,895 |
| Length | 95 |
| Max. P | 0.754617 |

| Location | 14,388,800 – 14,388,895 |
|---|---|
| Length | 95 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 80.20 |
| Shannon entropy | 0.37175 |
| G+C content | 0.40207 |
| Mean single sequence MFE | -27.72 |
| Consensus MFE | -13.99 |
| Energy contribution | -14.48 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.20 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.59 |
| SVM RNA-class probability | 0.754617 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
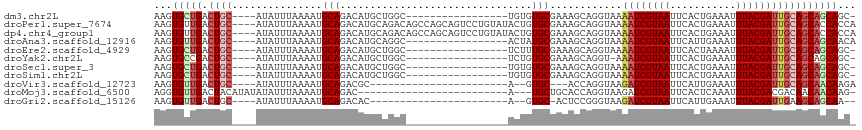
>dm3.chr2L 14388800 95 - 23011544 AAGUGCUGACUGC----AUAUUUAAAAUGCAGACAUGCUGGC-----------------UGUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCAGCAGC- ....(((..((((----((.......))))))....((((.(-----------------(((.(((....))).....((((((((...........)))))))))))))))))))- ( -31.60, z-score = -2.44, R) >droPer1.super_7674 306 113 - 1458 AAGUGUUGACUGC----AUAUUUAAAAUGCAGACAUGCAGACAGCCAGCAGUCCUGUAUACUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCACCACCA ..((((((.((((----((.......))))))...(((((((........)).)))))((((.(((....))))))).((((((((...........)))))))).))))))..... ( -32.50, z-score = -2.43, R) >dp4.chr4_group1 1742952 113 + 5278887 AAGUGUUGACUGC----AUAUUUAAAAUGCAGACAUGCAGACAGCCAGCAGUCCUGUAUACUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCACCACCA ..((((((.((((----((.......))))))...(((((((........)).)))))((((.(((....))))))).((((((((...........)))))))).))))))..... ( -32.50, z-score = -2.43, R) >droAna3.scaffold_12916 14752413 96 + 16180835 AAGUGUUGACUGC----AUAUUUAAAAUGCAGACAUGCAGGC-----------------ACUAUGCGAAAGCAGGUAAAAUCGUAAUUCAUUGAAAUUUACGAUUGCAGCAGCAACA ...(((((.((((----..........((((....)))).((-----------------(((.(((....))))))..((((((((...........)))))))))).))))))))) ( -30.30, z-score = -3.55, R) >droEre2.scaffold_4929 6141797 95 - 26641161 AAGUGCUGACUGC----AUAUUUAAAAUGCAGACAUGCUGGC-----------------UCUUUGCGAAAGCAGGUAAAAUCGUAAUUCACUAAAAUUUACGAUUGCAGCAGCAGC- ....((((.((((----((.......))))))...(((((..-----------------..(((((....)))))...((((((((...........)))))))).))))).))))- ( -31.10, z-score = -3.01, R) >droYak2.chr2L 14492907 94 - 22324452 AAGUGCCGACUGC----AUAUUUAAAAUGCAGACAUGCUGGC-----------------UCUGUGCGAAAGCAGGU-AAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCAGCAGC- ...(((...((((----((.......))))))...(((((((-----------------(...(((....))))))-.((((((((...........)))))))).))))))))..- ( -28.90, z-score = -1.74, R) >droSec1.super_3 648288 95 - 7220098 AAGUGCUGACUGC----AUAUUUAAAAUGCAGACAUGCUGGC-----------------UGUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCAGCAGC- ....(((..((((----((.......))))))....((((.(-----------------(((.(((....))).....((((((((...........)))))))))))))))))))- ( -31.60, z-score = -2.44, R) >droSim1.chr2L 14137233 95 - 22036055 AAGUGCUGACUGC----AUAUUUAAAAUGCAGACAUGCUGGC-----------------UGUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCAGCAGC- ....(((..((((----((.......))))))....((((.(-----------------(((.(((....))).....((((((((...........)))))))))))))))))))- ( -31.60, z-score = -2.44, R) >droVir3.scaffold_12723 2996756 85 + 5802038 AAGUGUUGACUGC----AUAUUUAAAAUGCAGACGC-----------------------A--GUGC---ACCAGGUAAGAUCGUAAUUCAUUGAAAUUUACGAUUGCAGCAACAAGA ...(((((.((((----..........((((.....-----------------------.--.)))---)........((((((((...........)))))))))))))))))... ( -20.90, z-score = -1.71, R) >droMoj3.scaffold_6500 16284303 88 + 32352404 AGGUGUUGACUACAUAUAUAUUUAAAAUGCAGAC-------------------------A---UGCUGCACCAGGUAAGAUCGUAAUUCACUCAAAUUUACGACGACAACAACAAG- .(.(((((...................(((((..-------------------------.---..)))))..........((((((...........))))))...))))).)...- ( -14.80, z-score = -0.96, R) >droGri2.scaffold_15126 690172 85 - 8399593 AAGUGUUGACUGC----AUAUUUAAAAUGCAGACAC-----------------------A--GUGC-ACUCCGGGUAAGAUCGUAAUUCAUUGAAAUUUACGAUUGAAGCAGCAA-- ..((((...((((----((.......))))))))))-----------------------.--.(((-.(....)((..((((((((...........))))))))...)).))).-- ( -19.10, z-score = -1.01, R) >consensus AAGUGUUGACUGC____AUAUUUAAAAUGCAGACAUGCUGGC_________________ACUGUGCGAAAGCAGGUAAAAUCGUAAUUCACUGAAAUUUACGAUUGCAGCAGCAGC_ ...(((((.((((..................................................(((........))).((((((((...........)))))))))))))))))... (-13.99 = -14.48 + 0.50)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:55 2011