| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,375,091 – 14,375,181 |
| Length | 90 |
| Max. P | 0.913154 |

| Location | 14,375,091 – 14,375,181 |
|---|---|
| Length | 90 |
| Sequences | 8 |
| Columns | 96 |
| Reading direction | forward |
| Mean pairwise identity | 84.43 |
| Shannon entropy | 0.31218 |
| G+C content | 0.42004 |
| Mean single sequence MFE | -22.90 |
| Consensus MFE | -16.20 |
| Energy contribution | -17.17 |
| Covariance contribution | 0.97 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.23 |
| SVM RNA-class probability | 0.913154 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14375091 90 + 23011544 AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGUGGCGAGGCAAUCG-AGGGUAUGAAUGACAGA---- ......(((((((((.(((...............)))))))))))).((..((((((-.....))))))..)).-.................---- ( -22.26, z-score = -1.65, R) >droWil1.scaffold_180764 1347430 96 - 3949147 AUCAAACUUAAAUGCAUGCAUAAAUCAAUCAAUCGCACCAUUUGAUAGACAGAAAGACAGUCAAGGGAGACUCAUAUAGAGAGAAAUAUAUGUAGA ................((((((...............((.((((((.(.(.....).).))))))))...(((.....))).......)))))).. ( -11.20, z-score = 0.58, R) >droPer1.super_5 1667699 87 + 6813705 AAUCCAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGAGCCGAGACAGUCGGAGAGUCUGAAUGA-------- ..(((.(((((((((.(((...............)))))))))))).(((.((((((-.....)))))).))))))............-------- ( -29.66, z-score = -4.01, R) >droAna3.scaffold_12916 14737227 86 - 16180835 AACCAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGUGCCGAGACAGCCG-AGAGUAUGAAUGA-------- ......(((((((((.(((...............)))))))))))).....((((((-.....)))))).....-.............-------- ( -22.46, z-score = -2.49, R) >droEre2.scaffold_4929 6128539 86 + 26641161 AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGUGGCGAGGCAUUCG-AGGGUAUGAAUGA-------- ......(((((((((.(((...............))))))))))))....(.(((((-((((......))))))-))).)........-------- ( -24.96, z-score = -2.67, R) >droYak2.chr2L 14479288 86 + 22324452 AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCCCAGCAUUUGACAGACAGUCUCG-AGUGGCGAGGCAGUCG-AGGGUAUGAAUGA-------- ......(((((((((......................))))))))).(((.((((((-.....)))))).))).-.............-------- ( -20.95, z-score = -1.23, R) >droSec1.super_3 634802 90 + 7220098 AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGUGGCGAGGCGGUCG-AGGGUAUGAAUGACAGA---- ......(((((((((.(((...............)))))))))))).(((.((((((-.....)))))).))).-.................---- ( -25.86, z-score = -2.48, R) >droSim1.chr2L 14123772 90 + 22036055 AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG-AGUGGCGAGGCGGUCG-AGGGUAUGAAUGACAGA---- ......(((((((((.(((...............)))))))))))).(((.((((((-.....)))))).))).-.................---- ( -25.86, z-score = -2.48, R) >consensus AAACAAGUUAAAUGCAUGCAUAAAUCAAUCAAUCGCAGCAUUUGACAGACAGUCUCG_AGUGGCGAGGCAGUCG_AGGGUAUGAAUGA________ ......(((((((((.(((...............)))))))))))).....((((((......))))))........................... (-16.20 = -17.17 + 0.97)

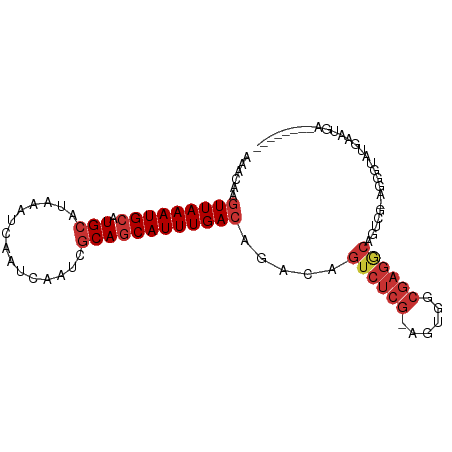
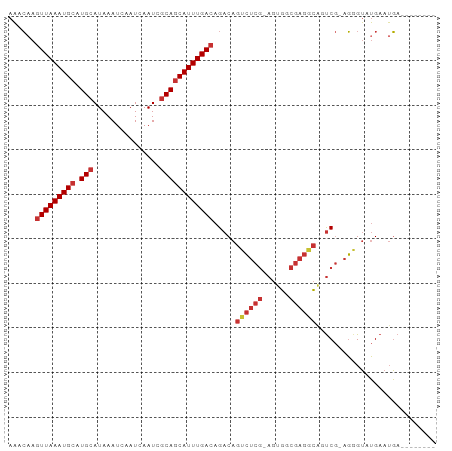
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:53 2011