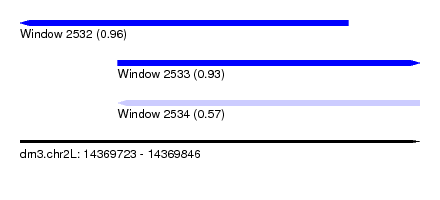
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,369,723 – 14,369,846 |
| Length | 123 |
| Max. P | 0.959633 |

| Location | 14,369,723 – 14,369,824 |
|---|---|
| Length | 101 |
| Sequences | 5 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 86.09 |
| Shannon entropy | 0.23615 |
| G+C content | 0.53538 |
| Mean single sequence MFE | -31.76 |
| Consensus MFE | -29.42 |
| Energy contribution | -28.98 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.77 |
| Structure conservation index | 0.93 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.67 |
| SVM RNA-class probability | 0.959633 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
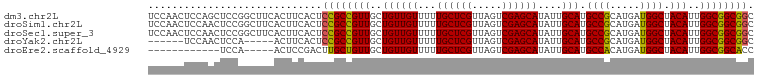
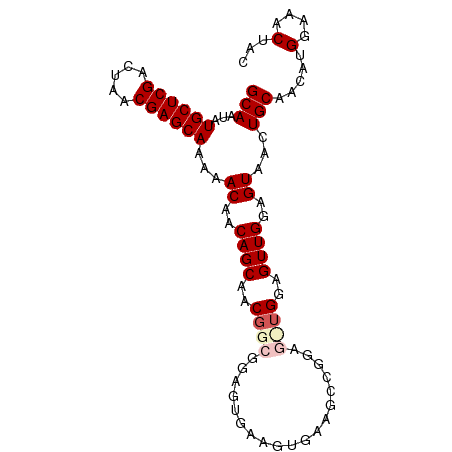
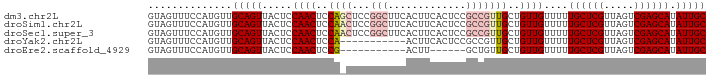
>dm3.chr2L 14369723 101 - 23011544 UCCAACUCCAGCUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGC ...(((..((((..((((.............))))..))))..)))..((((((.....))))))....((.((((((.(((((....))))))))))))) ( -37.62, z-score = -2.60, R) >droSim1.chr2L 14118310 101 - 22036055 UCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGC ..((((..((((..(((............)))..))))..))))....((((((.....))))))....((.((((((.(((((....))))))))))))) ( -32.80, z-score = -1.61, R) >droSec1.super_3 629341 101 - 7220098 UCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGC ..((((..((((..(((............)))..))))..))))....((((((.....))))))....((.((((((.(((((....))))))))))))) ( -32.80, z-score = -1.61, R) >droYak2.chr2L 14473766 90 - 22324452 ------UCCAACUCCA-----ACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGC ------..........-----........((((((((..((((((...((((((.....))))))....))).((((.....)))).)))..)))))))). ( -30.20, z-score = -1.69, R) >droEre2.scaffold_4929 6123401 84 - 26641161 ------------UCCA-----ACUCCGACUUGCUGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCACAUGAUGGCUACAUUGGCGGCACC ------------..((-----((..((((.....))))..))))....((((((.....))))))...(((.(((((.(((.......))))))))))).. ( -25.40, z-score = -1.32, R) >consensus UCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGCAUGCCGCAUGAUGGCUACAUUGGCGGCGGC .............................((((((((..((((((...((((((.....))))))....))).((((.....)))).)))..)))))))). (-29.42 = -28.98 + -0.44)
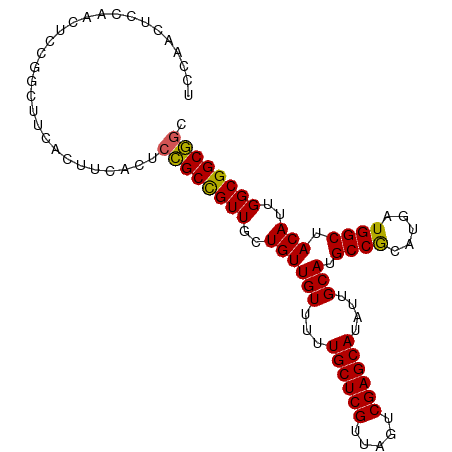
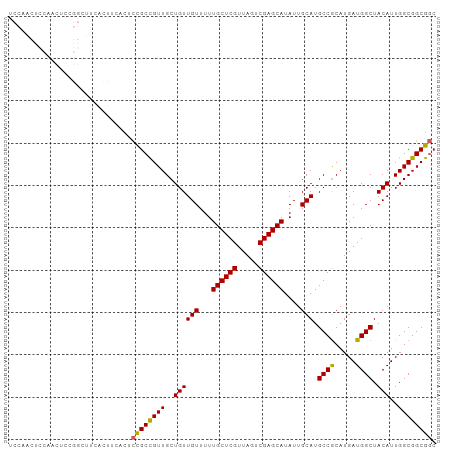
| Location | 14,369,753 – 14,369,846 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 89.12 |
| Shannon entropy | 0.18287 |
| G+C content | 0.47463 |
| Mean single sequence MFE | -24.11 |
| Consensus MFE | -17.63 |
| Energy contribution | -18.55 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.05 |
| Mean z-score | -2.62 |
| Structure conservation index | 0.73 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.929744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14369753 93 + 23011544 GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGCUGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....))))))..(((..((((..((((.............))))..))))..)))........))).....(....)... ( -28.82, z-score = -3.33, R) >droSim1.chr2L 14118340 93 + 22036055 GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGUUGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....))))))..(((..((((..((((.............))))..))))..)))........))).....(....)... ( -26.42, z-score = -2.63, R) >droSec1.super_3 629371 93 + 7220098 GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAGUUGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....))))))..(((..((((..((((.............))))..))))..)))........))).....(....)... ( -26.42, z-score = -2.63, R) >droYak2.chr2L 14473796 82 + 22324452 GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGU-----------UGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....))))))...((..((((..((((........))-----------))..))))..))...))).....(....)... ( -22.00, z-score = -2.69, R) >droEre2.scaffold_4929 6123431 76 + 26641161 GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACAGC------AAGU-----------CGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....)))))).........((..((((------....-----------....))))..))...))).....(....)... ( -16.90, z-score = -1.79, R) >consensus GCAAUAUGCUCGACUAACGAGCAAAAACAACAGCAACGGCGGAGUGAAGUGAAGCCGGAG_UGGAGUUGGAGUAACUGCAACAUGGAAACUAC (((...((((((.....))))))...((..((((..((((...................))))..))))..))...))).....(....)... (-17.63 = -18.55 + 0.92)

| Location | 14,369,753 – 14,369,846 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 93 |
| Reading direction | reverse |
| Mean pairwise identity | 89.12 |
| Shannon entropy | 0.18287 |
| G+C content | 0.47463 |
| Mean single sequence MFE | -20.42 |
| Consensus MFE | -14.40 |
| Energy contribution | -15.86 |
| Covariance contribution | 1.46 |
| Combinations/Pair | 1.09 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.571519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
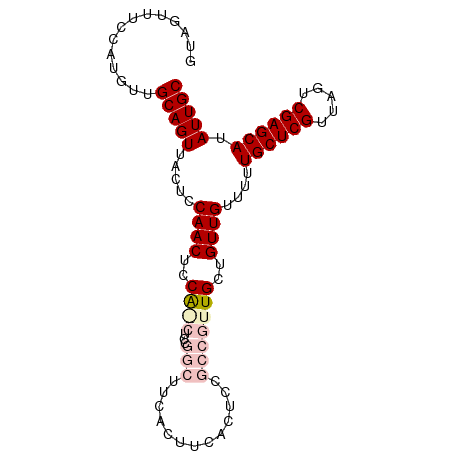
>dm3.chr2L 14369753 93 - 23011544 GUAGUUUCCAUGUUGCAGUUACUCCAACUCCAGCUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((......(((..((((..((((.............))))..))))..)))..((((((.....)))))).))))) ( -25.42, z-score = -3.16, R) >droSim1.chr2L 14118340 93 - 22036055 GUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((.....((((..((((..(((............)))..))))..))))....((((((.....)))))).))))) ( -20.60, z-score = -1.84, R) >droSec1.super_3 629371 93 - 7220098 GUAGUUUCCAUGUUGCAGUUACUCCAACUCCAACUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((.....((((..((((..(((............)))..))))..))))....((((((.....)))))).))))) ( -20.60, z-score = -1.84, R) >droYak2.chr2L 14473796 82 - 22324452 GUAGUUUCCAUGUUGCAGUUACUCCAACUCCA-----------ACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((.....((((..((-----------((...........))))..))))....((((((.....)))))).))))) ( -17.10, z-score = -1.71, R) >droEre2.scaffold_4929 6123431 76 - 26641161 GUAGUUUCCAUGUUGCAGUUACUCCAACUCCG-----------ACUU------GCUGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((.....((((..((-----------((..------...))))..))))....((((((.....)))))).))))) ( -18.40, z-score = -1.69, R) >consensus GUAGUUUCCAUGUUGCAGUUACUCCAACUCCA_CUCCGGCUUCACUUCACUCCGCCGUUGCUGUUGUUUUUGCUCGUUAGUCGAGCAUAUUGC ..............(((((.....((((..((((..(((............)))..))))..))))....((((((.....)))))).))))) (-14.40 = -15.86 + 1.46)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:52 2011