| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,271,254 – 14,271,346 |
| Length | 92 |
| Max. P | 0.599722 |

| Location | 14,271,254 – 14,271,346 |
|---|---|
| Length | 92 |
| Sequences | 9 |
| Columns | 103 |
| Reading direction | reverse |
| Mean pairwise identity | 76.28 |
| Shannon entropy | 0.45479 |
| G+C content | 0.61882 |
| Mean single sequence MFE | -35.35 |
| Consensus MFE | -17.21 |
| Energy contribution | -17.01 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.17 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.49 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.22 |
| SVM RNA-class probability | 0.599722 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14271254 92 - 23011544 AAAAGGCGCCGACCGAGUGUUCAAAUCACU--GGGAAGUCAACCCACGAUCUCGCUGCAGGC---------GCCAUUUGAGCCAUUUGAUGGCGCCGCCGCCG ....(((((.((.((((((.......))))--(((.......))).)).))..)).((.(((---------((((((.((....)).))))))))))).))). ( -34.00, z-score = -1.63, R) >droSim1.chr2L 14014292 92 - 22036055 AAAAGGCGCCGACCAAGUGUUCAAAUCACU--GGGAAGUCAACCCACGAUCUCGCUGCAGGC---------GCCAUUUGAGCCAUUUGAUGGCGCCGCCACCG ....(((((.(.(((.(((.......))))--))((..((.......))..)).).)).(((---------((((((.((....)).)))))))))))).... ( -32.30, z-score = -1.90, R) >droSec1.super_3 528730 92 - 7220098 AAAAGGCGCCGACCAAGUGUUCAAAUCACU--GGGAAGUCAACCCACGAUCUCGCUGCAGGC---------GCCAUUUGAGCCAUUUGAUGGCGCCGCCGCCG ....(((((...(((.(((.......))))--))((..((.......))..)))).((.(((---------((((((.((....)).))))))))))).))). ( -32.90, z-score = -1.59, R) >droYak2.chr2L 14371005 92 - 22324452 AAAAGGCGCCGACCGAGUGCUCAAAUCGCG--GGGAAGUCAACCCACGAUCGCGCUGCAGGC---------GCCAUUUGAGCCAUUUGAUGGCGCCGCCGCCG ....(((((((..(((((((((((((((.(--((........))).)))..((((.....))---------))..)))))).)))))).)))))))....... ( -42.20, z-score = -2.45, R) >droEre2.scaffold_4929 15446170 101 + 26641161 AAAAGGCGCCGACCGAGUGCUCAAAUCACG--GGGAAGUCAACCCACGAUCGCGCUGCAGGCGCCAUUUCAGCCAUUUGAGCCAUUUGAUGGCGCCGCCGCCG ....(((((((..(((((((((((((..((--((........)))..((..((((.....))))....)).)..))))))).)))))).)))))))....... ( -39.60, z-score = -1.71, R) >droAna3.scaffold_12916 4667728 81 + 16180835 AAAAGGCGCCGACCAAG--------CCGCU--GGGAAGUCAGCCCACGAUCGCGCUGCAGGC---------GCCAUUUGAACCAUUUGAUGGCGCCGCCG--- ....((((((((.....--------..(((--((....)))))......))).))....(((---------((((((..........)))))))))))).--- ( -31.52, z-score = -1.03, R) >droVir3.scaffold_12723 1468382 80 - 5802038 AAAAGGCGCCGCCAG---AGUCAGCC------ACGGUGUCAGCCCACGA--AUGCUUCAGGC---------GCCGCUUG-GCCAUUUGAUGGCGCCGCCAA-- ....(((((((.(((---(....(((------(((((((((((......--..)))...)))---------))))..))-))..)))).))))))).....-- ( -35.50, z-score = -2.07, R) >droMoj3.scaffold_6500 22459854 80 + 32352404 AAAAGGCGCCGCCAG---UUGCGAGC------CUGGUGUCAGCCCACGA--AUGCUUCAGGC---------GCCGCUUG-GCCAUUUGAUGGCGCCGCCAU-- ....(((((((((((---..(((.((------((((....(((......--..)))))))))---------..))))))-))(....)..)))))).....-- ( -33.30, z-score = -0.83, R) >droGri2.scaffold_15252 4831935 88 - 17193109 AAAAGGCGCCGCCGGAGCAGGCAAGCAGUCAAUCGGUGUCAGCCCACGA--UAGCUCCAGGC---------GCCGCUUA-GCCAUUUGAUGGCGCCGCCA--- ....((((.(((((((((.(((.....))).((((.((......)))))--).))))).(((---------........-))).......)))).)))).--- ( -36.80, z-score = -1.51, R) >consensus AAAAGGCGCCGACCGAGUGUUCAAAUCACU__GGGAAGUCAACCCACGAUCUCGCUGCAGGC_________GCCAUUUGAGCCAUUUGAUGGCGCCGCCGCCG ....((((((((........))...............(((((....((....)).....(((.........)))...........)))))))))))....... (-17.21 = -17.01 + -0.20)

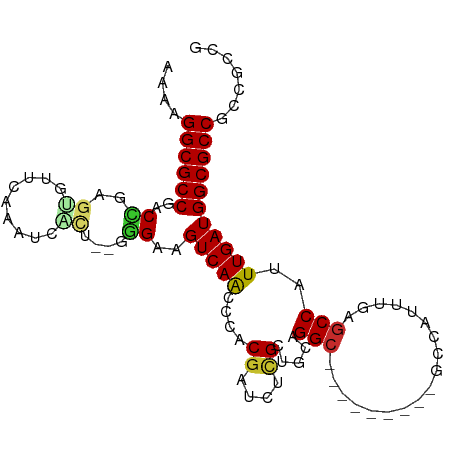

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:48 2011