| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,270,917 – 14,271,024 |
| Length | 107 |
| Max. P | 0.977234 |

| Location | 14,270,917 – 14,271,024 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 74.33 |
| Shannon entropy | 0.52852 |
| G+C content | 0.40469 |
| Mean single sequence MFE | -19.57 |
| Consensus MFE | -10.48 |
| Energy contribution | -10.78 |
| Covariance contribution | 0.30 |
| Combinations/Pair | 1.33 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.614739 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
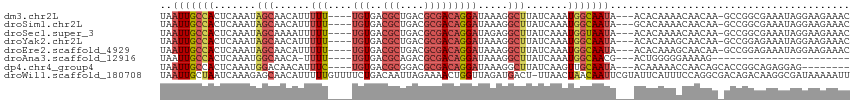
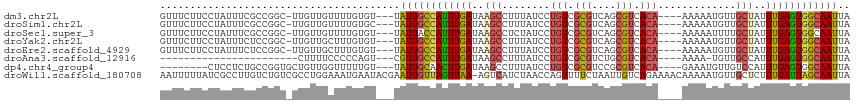
>dm3.chr2L 14270917 107 + 23011544 UAAUUGCCACUCAAAUAGCAACAUUUUU----UGUGACGCUGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAAUA---ACACAAAACAACAA-GCCGGCGAAAUAGGAAGAAAC ..(((((((.......(((....(((.(----(.((.(((....))).)).)).))).))).......))))))).---..............-.((.........))....... ( -18.84, z-score = -0.95, R) >droSim1.chr2L 14013958 107 + 22036055 UAAUUGCCACUCAAAUAGCAACAUUUUU----UGUGACGCUGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAAUA---GCACAAAACAACAA-GCCGGCGAAAUAGGAAGAAAC ...(((((...............(((.(----(.((.(((....))).)).)).)))(((((......((((....---)).)).......))-))))))))............. ( -20.72, z-score = -1.10, R) >droSec1.super_3 528396 107 + 7220098 UAAUUGCCACUCAAAUAGCAAAAUUUUU----UGUGACGCUGACGCGACAGGAUAGAGGCUUAUCAAAUGGUAAUA---ACACAAAACAACAA-GCCGGCGAAAUAGGAAGAAAC ...(((((...............(((.(----(.((.(((....))).)).)).)))(((((........((....---..))........))-))))))))............. ( -19.09, z-score = -0.99, R) >droYak2.chr2L 14370671 107 + 22324452 UAAUUGCCACUCAAAUAGCAACAUUUUU----UGUGACGCUGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAAUA---ACACAAAGCAACAA-GCCGGAGAAAUAGGAAGAAAC ..(((((((.......(((....(((.(----(.((.(((....))).)).)).))).))).......))))))).---..............-.((..(....).))....... ( -19.34, z-score = -1.07, R) >droEre2.scaffold_4929 15445836 107 - 26641161 UAAUUGCCACUCAAAUAGCAACAUUUUU----UGUGACGCUGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAAUA---ACACAAAGCAACAA-GCCGGAGAAAUAGGAAGAAAC ..(((((((.......(((....(((.(----(.((.(((....))).)).)).))).))).......))))))).---..............-.((..(....).))....... ( -19.34, z-score = -1.07, R) >droAna3.scaffold_12916 4667409 84 - 16180835 UAAUUGCCACUCAAAUGGCAACA-UUUU----UGUGACGCAGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAACG---ACUGGGGGAAAAG----------------------- .......(((..(((((....))-))).----.))).(((....))).((((((((....)))))...((....))---.))).........----------------------- ( -18.90, z-score = -1.55, R) >dp4.chr4_group4 1248879 100 - 6586962 UAAUUGCCACUCAAAUGGACAACAUUUC----UGUGACGCGGACGCGACAGGAUAAAGGCUUAUCAAGUUGCAAUA---ACAAAAACCAACAGCACCGGCAGAGGAG-------- ...(((((.......(((........((----((.....)))).(((((..(((((....)))))..)))))....---.......)))........))))).....-------- ( -21.56, z-score = -1.14, R) >droWil1.scaffold_180708 1160565 114 + 12563649 UAAUUGCUAAUCAAAGAGCAACAUUUUUGUUUUCUGACAAUUAGAAAACUGGUUAGAUGACU-UUAACUAACAAUUCGUAUUCAUUUCCAGGCGACAGACAAGGCGAUAAAAAUU ..((((((.........((.........(((((((((...)))))))))((((((((....)-))))))).....................)).........))))))....... ( -18.77, z-score = -0.68, R) >consensus UAAUUGCCACUCAAAUAGCAACAUUUUU____UGUGACGCUGACGCGACAGGAUAAAGGCUUAUCAAAUGGCAAUA___ACACAAAACAACAA_GCCGGCGAAAUAGGAAGAAAC ..(((((((............................(((....)))....(((((....)))))...)))))))........................................ (-10.48 = -10.78 + 0.30)
| Location | 14,270,917 – 14,271,024 |
|---|---|
| Length | 107 |
| Sequences | 8 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 74.33 |
| Shannon entropy | 0.52852 |
| G+C content | 0.40469 |
| Mean single sequence MFE | -23.14 |
| Consensus MFE | -13.91 |
| Energy contribution | -14.02 |
| Covariance contribution | 0.11 |
| Combinations/Pair | 1.35 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.60 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.97 |
| SVM RNA-class probability | 0.977234 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
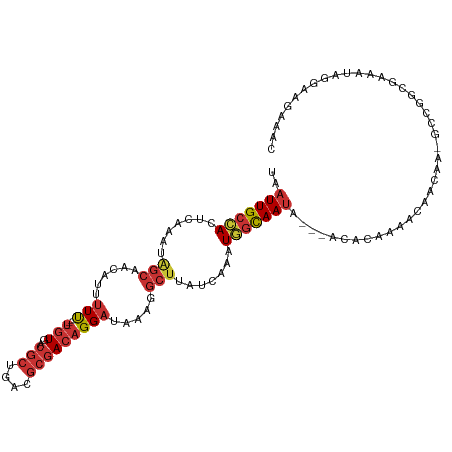
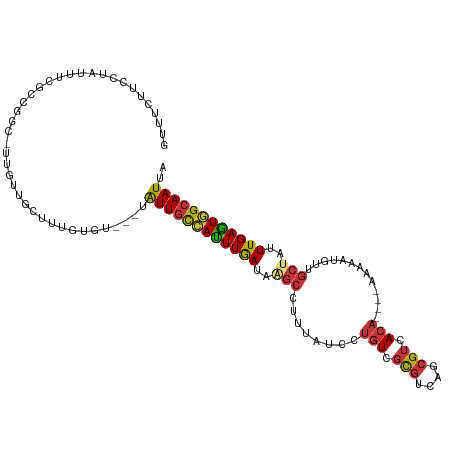
>dm3.chr2L 14270917 107 - 23011544 GUUUCUUCCUAUUUCGCCGGC-UUGUUGUUUUGUGU---UAUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCAGCGUCACA----AAAAAUGUUGCUAUUUGAGUGGCAAUUA ..................(((-((((((...((.((---....))))..))))))))).......((.(((....))).))..----......((((((((....)))))))).. ( -24.00, z-score = -1.60, R) >droSim1.chr2L 14013958 107 - 22036055 GUUUCUUCCUAUUUCGCCGGC-UUGUUGUUUUGUGC---UAUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCAGCGUCACA----AAAAAUGUUGCUAUUUGAGUGGCAAUUA ..................(((-((((((...((.((---....))))..))))))))).......((.(((....))).))..----......((((((((....)))))))).. ( -26.10, z-score = -2.00, R) >droSec1.super_3 528396 107 - 7220098 GUUUCUUCCUAUUUCGCCGGC-UUGUUGUUUUGUGU---UAUUACCAUUUGAUAAGCCUCUAUCCUGUCGCGUCAGCGUCACA----AAAAAUUUUGCUAUUUGAGUGGCAAUUA ..................(((-((((((..(.(((.---...))).)..))))))))).......((.(((....))).))..----.......(((((((....)))))))... ( -23.10, z-score = -1.92, R) >droYak2.chr2L 14370671 107 - 22324452 GUUUCUUCCUAUUUCUCCGGC-UUGUUGCUUUGUGU---UAUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCAGCGUCACA----AAAAAUGUUGCUAUUUGAGUGGCAAUUA ..................(((-((((((...((.((---....))))..))))))))).......((.(((....))).))..----......((((((((....)))))))).. ( -23.30, z-score = -1.66, R) >droEre2.scaffold_4929 15445836 107 + 26641161 GUUUCUUCCUAUUUCUCCGGC-UUGUUGCUUUGUGU---UAUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCAGCGUCACA----AAAAAUGUUGCUAUUUGAGUGGCAAUUA ..................(((-((((((...((.((---....))))..))))))))).......((.(((....))).))..----......((((((((....)))))))).. ( -23.30, z-score = -1.66, R) >droAna3.scaffold_12916 4667409 84 + 16180835 -----------------------CUUUUCCCCCAGU---CGUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCUGCGUCACA----AAAA-UGUUGCCAUUUGAGUGGCAAUUA -----------------------.............---.(((((((((..(...((........(((.(((....))).)))----....-....))...)..))))))))).. ( -19.79, z-score = -2.56, R) >dp4.chr4_group4 1248879 100 + 6586962 --------CUCCUCUGCCGGUGCUGUUGGUUUUUGU---UAUUGCAACUUGAUAAGCCUUUAUCCUGUCGCGUCCGCGUCACA----GAAAUGUUGUCCAUUUGAGUGGCAAUUA --------......(((((((..(((..(...((((---....)))).)..))).)))......((((.(((....))).)))----)(((((.....)))))....)))).... ( -21.60, z-score = -0.13, R) >droWil1.scaffold_180708 1160565 114 - 12563649 AAUUUUUAUCGCCUUGUCUGUCGCCUGGAAAUGAAUACGAAUUGUUAGUUAA-AGUCAUCUAACCAGUUUUCUAAUUGUCAGAAAACAAAAAUGUUGCUCUUUGAUUAGCAAUUA ....(((((..((..((.....))..))..)))))....(((((((((((((-((.((.(......(((((((.......)))))))......).))..))))))))))))))). ( -23.90, z-score = -2.36, R) >consensus GUUUCUUCCUAUUUCGCCGGC_UUGUUGCUUUGUGU___UAUUGCCAUUUGAUAAGCCUUUAUCCUGUCGCGUCAGCGUCACA____AAAAAUGUUGCUAUUUGAGUGGCAAUUA ........................................(((((((((((((((....))))).((.(((....))).))......................)))))))))).. (-13.91 = -14.02 + 0.11)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:47 2011