
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,244,248 – 14,244,357 |
| Length | 109 |
| Max. P | 0.545908 |

| Location | 14,244,248 – 14,244,357 |
|---|---|
| Length | 109 |
| Sequences | 11 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 69.19 |
| Shannon entropy | 0.58982 |
| G+C content | 0.51889 |
| Mean single sequence MFE | -28.41 |
| Consensus MFE | -15.06 |
| Energy contribution | -14.99 |
| Covariance contribution | -0.07 |
| Combinations/Pair | 1.07 |
| Mean z-score | -0.74 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.545908 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14244248 109 + 23011544 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUCAUGA---AGUUUCUCUGCCAGCAGCAGCAUCCACA-----GCUGUUCGCCAAAAAACAAAAAACAAAAAACCCGAAGGA ((((((((((....))))))....(((((((.(((....)))---.))))).))))))((((((((.......-----)))))).)).....................((....)). ( -27.10, z-score = -0.94, R) >droSim1.chr2L 13987444 103 + 22036055 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUCAUGA---AGUUUCUCUGCCAGCAGCAGCAUCCACA-----GCUGUUCGCCAAAAAAAAAAAAAC------CCCGAAGGA ((((((((((....))))))....(((((((.(((....)))---.))))).))))))((((((((.......-----)))))).))..............(------(.....)). ( -27.10, z-score = -0.96, R) >droSec1.super_3 501723 103 + 7220098 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUCAUGA---AGUUUCUCUGCCAGCAGCAGCAUCCACA-----GCUGUUCGCCAAAAAAAAAAAAAC------CCCGAAGGA ((((((((((....))))))....(((((((.(((....)))---.))))).))))))((((((((.......-----)))))).))..............(------(.....)). ( -27.10, z-score = -0.96, R) >droYak2.chr2L 14342661 95 + 22324452 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUCAUGA---AGUUACCCUGCCAGCAGCAGCAUCCACA-----GCUGUUCGCCAAGAAAGCCGAAGGA-------------- ...(((((((...(((((.((....((.(((.(((....)))---.))).))..)))))))(((((.......-----)))))...........)))).))).-------------- ( -27.80, z-score = -0.63, R) >droEre2.scaffold_4929 15419028 94 - 26641161 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUCAUGA---AGUUUCCCUGCCAGCAGCAGCAUCCACA-----GCUGUUCGCCAA-AAAUCCGAAGGA-------------- ((((((((((....)))))).....((((((.(((....)))---.))))))..))))((((((((.......-----)))))).))...-............-------------- ( -29.00, z-score = -1.45, R) >droAna3.scaffold_12916 4641864 88 - 16180835 UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACCGA---------AGUCGUCCUGACAGCAGCAUCCCUA---CAGCUGUUGUC--CAAGUGCGAAGGA--------------- ((..((((((....))))))..)).(....)....((..---------..(((..(((((((((((.......---..)))))))))--..))..))).)).--------------- ( -31.80, z-score = -2.20, R) >dp4.chr4_group4 1219538 103 - 6586962 UGGCCUCCACAAAUGUUGAGUUCAAGGAGACGUCACCGAUGG------AGAUGUCGCGGUAGCAGCAUCCACAUAACAGCUGUUGGC--CAAGAGCGAGAGACACCAGCGC------ (((((...(((..(((((.((....(((((((((.((...))------.))))))((....))....))))).)))))..))).)))--))...((...........))..------ ( -29.30, z-score = -0.36, R) >droPer1.super_16 1584796 103 + 1990440 UGGCCUCCACAAAUGUUGAGUUCAAGGAGACGUCACCGAUGG------AGAUGUCGCGGUAGCAGCAUCCACAUAACAGCUGUUGGC--CAAGAGCGAGAGACACCAGUGC------ .......(((...((((..((((.....((((((.((...))------.))))))..(((((((((............)))))).))--)..))))....))))...))).------ ( -28.60, z-score = -0.17, R) >droVir3.scaffold_12723 1429201 95 + 5802038 UGGCCUCGACAAGUGUUGAGUUCAAGGAGACGUCACUGUUGA---GGUCCGCGUCCACGUUGCAGCUUCCCCAUA-CAGCUGUUGUC---CAGAGCGAAGGA--------------- .((((((((((.((((((....)))(....)..)))))))))---))))(((.((......((((((........-.))))))....---..))))).....--------------- ( -31.10, z-score = -0.77, R) >droMoj3.scaffold_6500 22417827 98 - 32352404 UGGCCUCGACAAAUGUUGAGUUCAAGGAGACGUCACUGUUCA---GGUACUCGCAACUAUUGCAGCUUCCCCACU-CAGCUGUUGUUAGUCAGAGCGAAGGA--------------- ((..((((((....))))))..)).(....)((.(((.....---)))))((((.((((..((((((........-.))))))...))))....))))....--------------- ( -25.70, z-score = 0.40, R) >droGri2.scaffold_15252 4797881 96 + 17193109 UGGCCUCGACAAAUGUUGAGUUCAAGGAGACGUCACUGAUGAUGAGGUGUUCUCUGCAGCUGCAGCUUCCCCA---CAGCUGUUAUC---CAAAGCGAAGGA--------------- ((..((((((....))))))..)).(((((...((((........)))).)))))(((((((...........---)))))))..((---(........)))--------------- ( -27.90, z-score = -0.07, R) >consensus UGGCCUCGGCAAAUGUUGAGUUCAAGGAGACGUCACUGAUGA___AGUUUCCCUGCCAGCAGCAGCAUCCACA___CAGCUGUUCGC__AAAAAGCGAAAGA_______________ ((..((((((....))))))..)).(....)..............................(((((............))))).................................. (-15.06 = -14.99 + -0.07)

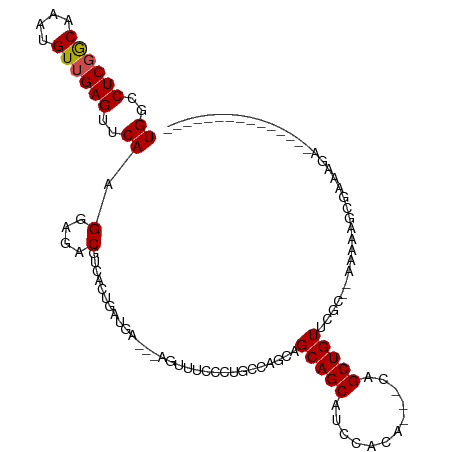
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:45 2011