| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,243,274 – 14,243,405 |
| Length | 131 |
| Max. P | 0.994521 |

| Location | 14,243,274 – 14,243,375 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 105 |
| Reading direction | forward |
| Mean pairwise identity | 87.35 |
| Shannon entropy | 0.23977 |
| G+C content | 0.53254 |
| Mean single sequence MFE | -37.10 |
| Consensus MFE | -26.30 |
| Energy contribution | -27.75 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.08 |
| Structure conservation index | 0.71 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.34 |
| SVM RNA-class probability | 0.988852 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
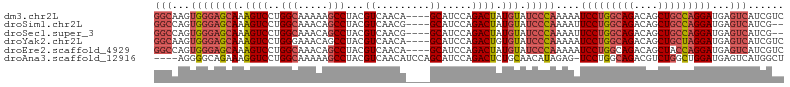
>dm3.chr2L 14243274 101 + 23011544 GGCAAGUGGGAGCAAAGUCCUGGCAAAAAGCCUACGUCAACA----GCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUC (((...((((((((.((((..(((.....)))...((.....----)).....)))).))).)))))...((((((((((....))))))))))..)))...... ( -38.40, z-score = -4.22, R) >droSim1.chr2L 13986486 99 + 22036055 GGCCAGUGGGAGCAAAGUCCUGGCAAACAGCCUACGUCAACG----GCAUCCAGACUAUGUAUCCCAAAAUUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCG-- (((((.((((((((.((((.(((......(((.........)----))..))))))).))).)))))....(((((((((....))))))))))).)))....-- ( -39.20, z-score = -3.72, R) >droSec1.super_3 500765 99 + 7220098 GGCCAGUGGGAGCAAAGUCCUGGCAAACAGCCUACGUCAACG----GCAUCCAGACUAUGUAUCCCAAAAUUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCG-- (((((.((((((((.((((.(((......(((.........)----))..))))))).))).)))))....(((((((((....))))))))))).)))....-- ( -39.20, z-score = -3.72, R) >droYak2.chr2L 14341687 101 + 22324452 GGCAAGUGGGAGCAAAGUCCUGGGAAACAGCCUACGUCAACA----GCAUCCAGACUGUGUAUCCCAAAAAUCCUGGCAGACAGCUGCUAGGAUGAGUCAUCGUC (((...((((((((.((((.(((....).((...........----))...)))))).))).)))))...((((((((((....))))))))))..)))...... ( -35.40, z-score = -2.38, R) >droEre2.scaffold_4929 15418020 101 - 26641161 GGCCAGUGGGAGCAAAGUCCUGGCAAACAGCCUACGUCAACA----GCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUACCAGGAUGAGUCAUCGUC ((((..((((((((.((((..(((.....)))...((.....----)).....)))).))).)))))...(((((((.((....)).)))))))).)))...... ( -31.10, z-score = -2.03, R) >droAna3.scaffold_12916 4640859 100 - 16180835 ----AGGGGCAGAAAGGUCCUGGCAAAAAGCCUACGUCAACAUCCAGCAUCCAGACUCUGCAACAUAGAG-UCCUGGCAGACGUCUGGCUGGAUGAGUCAUGGCU ----.(((((......)))))(((.....)))...((((.((((((((..((((((((((.....)))))-).))))(((....))))))))))).....)))). ( -39.30, z-score = -2.43, R) >consensus GGCCAGUGGGAGCAAAGUCCUGGCAAACAGCCUACGUCAACA____GCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUC (((...((((((((.((((..(((.....)))...(....)............)))).))).)))))....(((((((((....)))))))))...)))...... (-26.30 = -27.75 + 1.45)
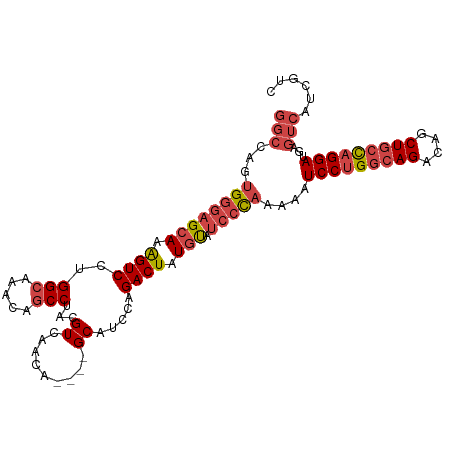
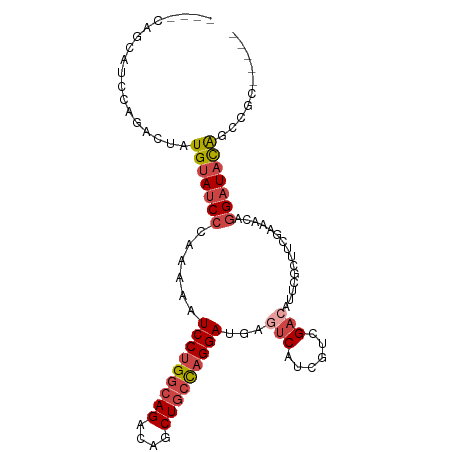
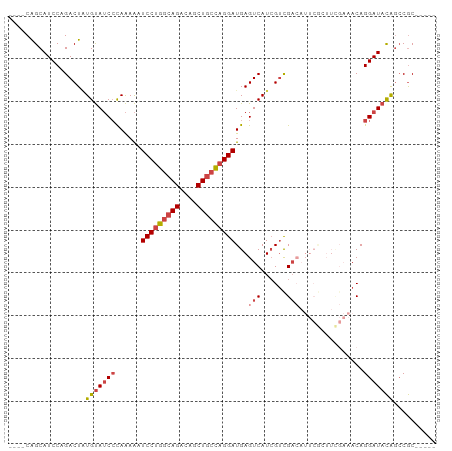
| Location | 14,243,314 – 14,243,405 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | forward |
| Mean pairwise identity | 79.76 |
| Shannon entropy | 0.35772 |
| G+C content | 0.51627 |
| Mean single sequence MFE | -28.65 |
| Consensus MFE | -20.13 |
| Energy contribution | -21.22 |
| Covariance contribution | 1.09 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.80 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.71 |
| SVM RNA-class probability | 0.994521 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14243314 91 + 23011544 ----CAGCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUCGACAUUCGCUUCGAAACAGGAUACAGCCGC----- ----..............(((((((.....((((((((((....))))))))))..(((......))).((((...))))...))))))).....----- ( -30.30, z-score = -3.51, R) >droSim1.chr2L 13986526 79 + 22036055 ----CGGCAUCCAGACUAUGUAUCCCAAAAUUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUCGA------------AACAGGAUACAGCCGC----- ----((((..........(((((((........(((((((....)))))))(((((....)))))..------------....))))))))))).----- ( -30.10, z-score = -4.11, R) >droSec1.super_3 500805 79 + 7220098 ----CGGCAUCCAGACUAUGUAUCCCAAAAUUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUCGA------------AACAGGAUACAGCCGC----- ----((((..........(((((((........(((((((....)))))))(((((....)))))..------------....))))))))))).----- ( -30.10, z-score = -4.11, R) >droYak2.chr2L 14341727 91 + 22324452 ----CAGCAUCCAGACUGUGUAUCCCAAAAAUCCUGGCAGACAGCUGCUAGGAUGAGUCAUCGUCGACAUUCGCUUCGAAACAGGAUACAGCCGC----- ----(((........)))(((((((.....((((((((((....))))))))))..(((......))).((((...))))...))))))).....----- ( -28.20, z-score = -2.19, R) >droEre2.scaffold_4929 15418060 91 - 26641161 ----CAGCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUACCAGGAUGAGUCAUCGUCGACAUUCGCUUCGAAACAGGAUACGGCCGC----- ----..............(((((((.....(((((((.((....)).)))))))..(((......))).((((...))))...))))))).....----- ( -23.00, z-score = -1.16, R) >droAna3.scaffold_12916 4640895 99 - 16180835 CAUCCAGCAUCCAGACUCUGCAACAUAGAG-UCCUGGCAGACGUCUGGCUGGAUGAGUCAUGGCUGCCAUUCAUUUUGAAACAGGAUAUACACACAAAGC ((((((((..((((((((((.....)))))-).))))(((....))))))))))).((.(((.(((...(((.....))).)))..)))))......... ( -30.20, z-score = -1.73, R) >consensus ____CAGCAUCCAGACUAUGUAUCCCAAAAAUCCUGGCAGACAGCUGCCAGGAUGAGUCAUCGUCGACAUUCGCUUCGAAACAGGAUACAGCCGC_____ ..................(((((((......(((((((((....)))))))))...(((......)))...............))))))).......... (-20.13 = -21.22 + 1.09)

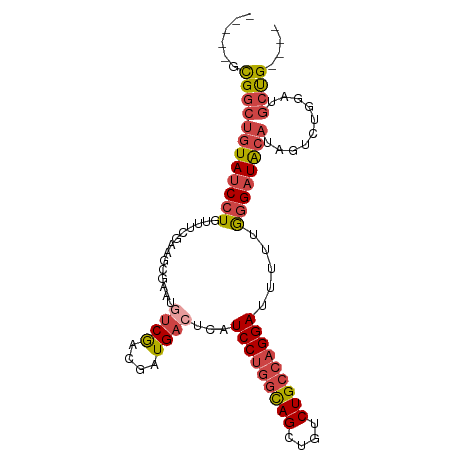
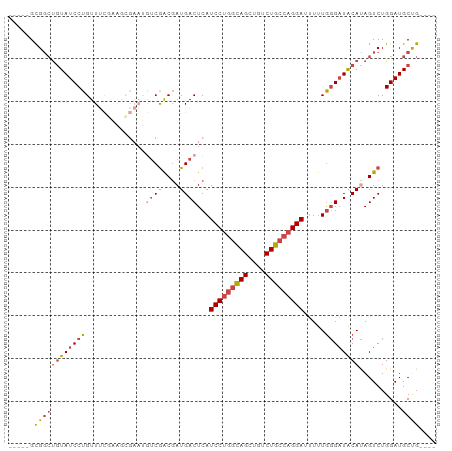
| Location | 14,243,314 – 14,243,405 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 100 |
| Reading direction | reverse |
| Mean pairwise identity | 79.76 |
| Shannon entropy | 0.35772 |
| G+C content | 0.51627 |
| Mean single sequence MFE | -30.15 |
| Consensus MFE | -21.23 |
| Energy contribution | -22.95 |
| Covariance contribution | 1.72 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.75 |
| Structure conservation index | 0.70 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.01 |
| SVM RNA-class probability | 0.873403 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14243314 91 - 23011544 -----GCGGCUGUAUCCUGUUUCGAAGCGAAUGUCGACGAUGACUCAUCCUGGCAGCUGUCUGCCAGGAUUUUUGGGAUACAUAGUCUGGAUGCUG---- -----..(((((((((((((((((...((.....)).))).)))..((((((((((....))))))))))....)))))))..)))).........---- ( -32.80, z-score = -2.19, R) >droSim1.chr2L 13986526 79 - 22036055 -----GCGGCUGUAUCCUGUU------------UCGACGAUGACUCAUCCUGGCAGCUGUCUGCCAGGAAUUUUGGGAUACAUAGUCUGGAUGCCG---- -----.(((((((((((((((------------........)))...(((((((((....))))))))).....))))))))..........))))---- ( -30.40, z-score = -2.76, R) >droSec1.super_3 500805 79 - 7220098 -----GCGGCUGUAUCCUGUU------------UCGACGAUGACUCAUCCUGGCAGCUGUCUGCCAGGAAUUUUGGGAUACAUAGUCUGGAUGCCG---- -----.(((((((((((((((------------........)))...(((((((((....))))))))).....))))))))..........))))---- ( -30.40, z-score = -2.76, R) >droYak2.chr2L 14341727 91 - 22324452 -----GCGGCUGUAUCCUGUUUCGAAGCGAAUGUCGACGAUGACUCAUCCUAGCAGCUGUCUGCCAGGAUUUUUGGGAUACACAGUCUGGAUGCUG---- -----..(((((((((((((((((...((.....)).))).)))..(((((.((((....)))).)))))....)))))..)))))).........---- ( -26.60, z-score = -0.27, R) >droEre2.scaffold_4929 15418060 91 + 26641161 -----GCGGCCGUAUCCUGUUUCGAAGCGAAUGUCGACGAUGACUCAUCCUGGUAGCUGUCUGCCAGGAUUUUUGGGAUACAUAGUCUGGAUGCUG---- -----(((.(((((((((((((((...((.....)).))).)))..((((((((((....))))))))))....))))))).......)).)))..---- ( -30.11, z-score = -1.44, R) >droAna3.scaffold_12916 4640895 99 + 16180835 GCUUUGUGUGUAUAUCCUGUUUCAAAAUGAAUGGCAGCCAUGACUCAUCCAGCCAGACGUCUGCCAGGA-CUCUAUGUUGCAGAGUCUGGAUGCUGGAUG ...(..((.((....((...(((.....))).))..))))..)..(((((((((((((.(((((...((-(.....)))))))))))))...)))))))) ( -30.60, z-score = -1.08, R) >consensus _____GCGGCUGUAUCCUGUUUCGAAGCGAAUGUCGACGAUGACUCAUCCUGGCAGCUGUCUGCCAGGAUUUUUGGGAUACAUAGUCUGGAUGCUG____ ...........((((((...(((.....)))....(((.(((.(((((((((((((....)))))))))....))))...))).))).))))))...... (-21.23 = -22.95 + 1.72)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:44 2011