| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,235,996 – 14,236,112 |
| Length | 116 |
| Max. P | 0.986209 |

| Location | 14,235,996 – 14,236,112 |
|---|---|
| Length | 116 |
| Sequences | 7 |
| Columns | 117 |
| Reading direction | forward |
| Mean pairwise identity | 84.53 |
| Shannon entropy | 0.30659 |
| G+C content | 0.42535 |
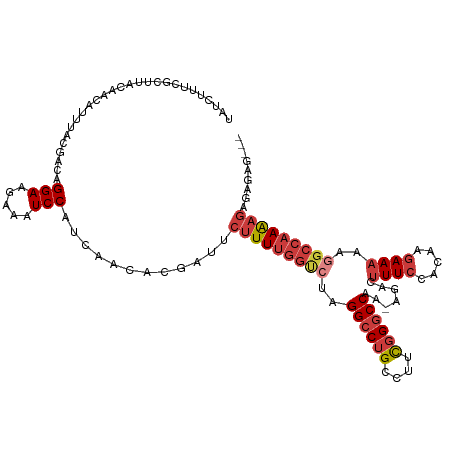
| Mean single sequence MFE | -30.73 |
| Consensus MFE | -18.94 |
| Energy contribution | -20.13 |
| Covariance contribution | 1.19 |
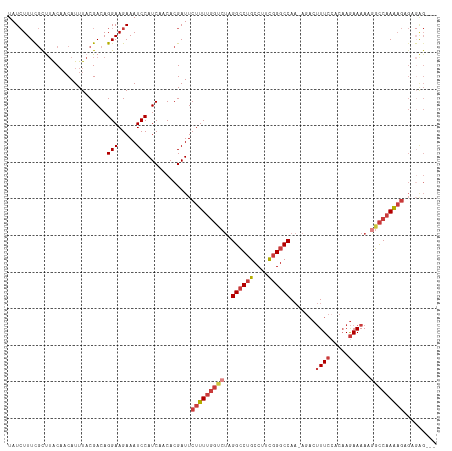
| Combinations/Pair | 1.14 |
| Mean z-score | -2.98 |
| Structure conservation index | 0.62 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.23 |
| SVM RNA-class probability | 0.986209 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
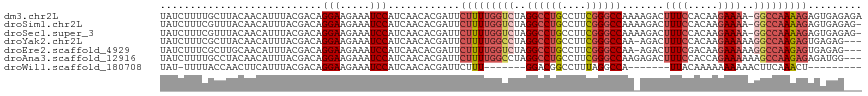
>dm3.chr2L 14235996 116 + 23011544 UAUCUUUUGCUUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGUCUAGGCCUGCCUUCGGGCCAAAAGACUUUCCACAAGAAAA-GGCCAAAAGAGUGAGAGA ..(((((..(.................(((.....)))...........(((((((((((.((((((....)))))).......((((.....)))))-)))))))))))..))))) ( -34.60, z-score = -3.24, R) >droSim1.chr2L 13979202 115 + 22036055 UAUCUUUCGUUUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGUCUAGGCCUGCCUUCGGGCCAAAAGACUUUCCACAAGAAAA-GGCCAAAAGAGUGAGAG- ...(((((...................(((.....))).........(((((((((((((.((((((....)))))).......((((.....)))))-)))))))))))))))))- ( -33.40, z-score = -3.25, R) >droSec1.super_3 493479 115 + 7220098 UAUCUUUCGUUUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGUCUAGGCCUGCCUUCGGGCCAAAAGACUUUCCACAAGAAAA-GGCCAAAAGAGUGAGAG- ...(((((...................(((.....))).........(((((((((((((.((((((....)))))).......((((.....)))))-)))))))))))))))))- ( -33.40, z-score = -3.25, R) >droYak2.chr2L 14334464 113 + 22324452 UAUCUUUCGCUUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGCCUAGGCCUGCCUUCGGGCCAA-AGACUUUCCACAAGAAAAAGGCCAAGAGUGAGAG--- ...(((((((((...............(((.....)))...............(((((((.((((((....))))))..-....((((.....)))).))))))))))))))))--- ( -33.80, z-score = -3.38, R) >droEre2.scaffold_4929 15410810 113 - 26641161 UAUCUUUCGCUUGCAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGUCUAGGCCUGCCUUCGGGCCAA-AGACUUUCGACAAGAAAAAGGCCAAGAGUGAGAG--- ...(((((((((...........(((..((((((.((.........))))))))..)))..((((((((....)))...-....((((.....)))).)))))..)))))))))--- ( -35.40, z-score = -3.06, R) >droAna3.scaffold_12916 4633670 114 - 16180835 UAUCUUUUGCCUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGCCUAGGCCUGCCUUCGGGCCAAGAGACUUUCCACCAGAAAAAAGCCAAGAGAGAUGG--- ..(((((((.((...............(((.....)))...........((((((((((((((....)))..)))))))))))................)).))))))).....--- ( -30.00, z-score = -2.20, R) >droWil1.scaffold_180708 1219626 93 + 12563649 UAU-UUUUACCAACUUCAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUU-------GGACGGCCUUUAGGCCA-------UUACAAAAAAAAAACUUCAAACU--------- ...-........................((((...((((..............)-------))).((((....)))).-------..............)))).....--------- ( -14.54, z-score = -2.47, R) >consensus UAUCUUUCGCUUACAACAUUUACGACAGGAAGAAAUCCAUCAACACGAUUCUUUUGGUCUAGGCCUGCCUUCGGGCCAA_AGACUUUCCACAAGAAAAAGGCCAAAAGAGAGAG___ ...........................(((.....)))............(((((((((..((((((....)))))).......((((.....))))..)))))))))......... (-18.94 = -20.13 + 1.19)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:41 2011