| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,230,537 – 14,230,648 |
| Length | 111 |
| Max. P | 0.953335 |

| Location | 14,230,537 – 14,230,648 |
|---|---|
| Length | 111 |
| Sequences | 7 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 58.40 |
| Shannon entropy | 0.84157 |
| G+C content | 0.38257 |
| Mean single sequence MFE | -25.04 |
| Consensus MFE | -10.26 |
| Energy contribution | -11.19 |
| Covariance contribution | 0.93 |
| Combinations/Pair | 1.78 |
| Mean z-score | -1.15 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.59 |
| SVM RNA-class probability | 0.953335 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
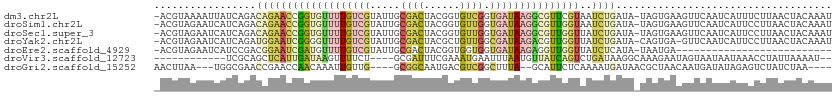
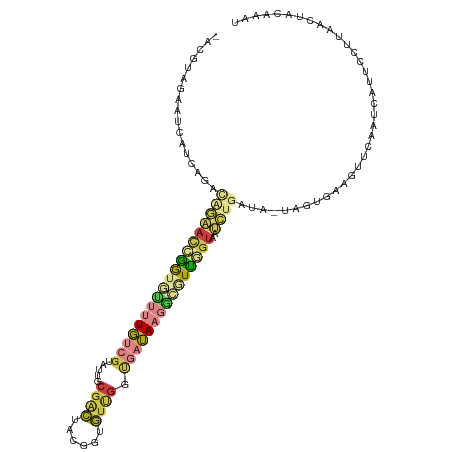
>dm3.chr2L 14230537 111 + 23011544 -ACGUAAAAUUAUCAGACAGAACCGGUGUUUUGUCGUAUUGCGACUACGGUGUCGGUGAUAAGGCGUUCGUAAUCUGAUA-UAGUGAAGUUCAAUCAUUUCUUAACUACAAAU -((((....((((((((((((((....))))))))......((((......)))).)))))).)))).............-(((((((((......)))))...))))..... ( -24.00, z-score = -0.20, R) >droSim1.chr2L 13973743 111 + 22036055 -ACGUAGAAUCAUCAGACAGAACCGGUGUUUUGUCGUAUUGCGACUACGGUGUUGGUGAUAAGGCGUUGGUAAUCUGAUA-UAGUGAAGUUCAAUCAUUCCUUAACUACAAAU -..((((....((((((....(((..(((((((((((...((.((....))))..)))))))))))..)))..)))))).-.(((((.......)))))......)))).... ( -29.20, z-score = -1.40, R) >droSec1.super_3 488060 111 + 7220098 -ACGUAGAAUCAUCAGACAGAACCGGUGUUUUGUCGUAUUGCGACUACGGUGUUGGUGAUAAGGCGUUGGUUAUCUGAUA-UAGUGAAGUUCAAUCAUUCCUUAACUACAAAU -..((((....((((((...((((..(((((((((((...((.((....))))..)))))))))))..)))).)))))).-.(((((.......)))))......)))).... ( -31.50, z-score = -2.07, R) >droYak2.chr2L 14328828 110 + 22324452 -ACGUAGAAUCAUCAGAUGGAAUCGGGGUUUUGUCGUAUUGCGACUACGCUGUUGGCGAUAAGACGUUGGUUAUCUGAUA-CAGUGA-GUUCAAUCAUUCCUUAACUACAAAU -..((((....((((((((..(((((.((((((((((...(((....))).....)))))))))).))))))))))))).-.(((((-......)))))......)))).... ( -35.20, z-score = -3.15, R) >droEre2.scaffold_4929 15405273 85 - 26641161 -ACGUAGAAUCAUCCGACGGAAUCGAUGUUUUGUCGUAUUGCGACUACGGUGGUGGUGAUAAGAGGUUGGUUAUCUCAUA-UAAUGA-------------------------- -.(((...(((((((((((((((....)))))))))....((.((....)).))))))))..(((((.....)))))...-..))).-------------------------- ( -22.60, z-score = -0.72, R) >droVir3.scaffold_12723 1406031 95 + 5802038 ------------UCGCAGCUCAUUGAUAAGUUUUCU----GCGAUUUCGAAAUGAAUUUAAUGUUAUCAGUCUGAUAAGGCAAAGAAUAGUAAUAAUAAACCUAUUAAAAU-- ------------((((((...(((....)))...))----))))......((((..((((.((((((...(((..........)))...)))))).))))..)))).....-- ( -13.20, z-score = 0.25, R) >droGri2.scaffold_15252 4777688 100 + 17193109 AACUUAA---UGGCGAACCGAACCAACAAAUUGUUG----GCGGCAAUGACGUCGGCUUUA--GCAUUCUCAAAAUGAUAACGCUAACAAUGAUAUAGAGUCUAUCUAA---- .......---(((((..(((..(((((.....))))----))))...(((.(...((....--))...)))).........))))).........((((.....)))).---- ( -19.60, z-score = -0.73, R) >consensus _ACGUAGAAUCAUCAGACAGAACCGGUGUUUUGUCGUAUUGCGACUACGGUGUUGGUGAUAAGGCGUUGGUAAUCUGAUA_UAGUGAAGUUCAAUCAUUCCUUAACUACAAAU .................(((((((((((((((((((.....((((......)))).)))))))))))))))..)))).................................... (-10.26 = -11.19 + 0.93)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:41 2011