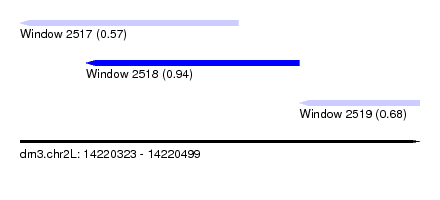
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,220,323 – 14,220,499 |
| Length | 176 |
| Max. P | 0.935947 |

| Location | 14,220,323 – 14,220,419 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 73.41 |
| Shannon entropy | 0.48932 |
| G+C content | 0.48616 |
| Mean single sequence MFE | -35.90 |
| Consensus MFE | -17.87 |
| Energy contribution | -18.32 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.36 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.50 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.16 |
| SVM RNA-class probability | 0.570293 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
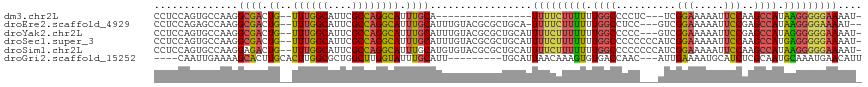
>dm3.chr2L 14220323 96 - 23011544 CCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCA----------------UUUUCUUUUUUGGCCCCUC---UCGGAAAAAUUCCAAGCCAUAAGGGGGAAAAU- (((((...((((((((((.((--((((((....)))))))).)))).----------------........)))))).....---..(((.....))).........))))).....- ( -31.60, z-score = -1.20, R) >droEre2.scaffold_4929 15395276 110 + 26641161 CCUCCAGAGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCA-UUUUCUUUUUUGGCCUCC---GUCGGAAAAAUUCCGAGCCAUAAGGGGAAAAU-- ....(((.((((((((((.((--((((((....)))))))).)))).))))...)).)))..-(((((((((.((((....---.(((((.....))))))))).)))))))))..-- ( -39.20, z-score = -2.19, R) >droYak2.chr2L 14318763 112 - 22324452 CCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCC---GUCGGAAAAAUUCCGAGCCAUAGGGGGGAAAAU- ....((((((((((((((.((--((((((....)))))))).)))).))))...))))))..((((((((((.((((....---.(((((.....))))))))).))))))))))..- ( -43.10, z-score = -2.35, R) >droSec1.super_3 477953 115 - 7220098 CCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCCCCAUCGGAAAAAUUCCAAGCCAUGAGGGGGAAAAU- ....((((((((((((((.((--((((((....)))))))).)))).))))...)))))).................(((((.(((.((............))))).))))).....- ( -39.40, z-score = -1.38, R) >droSim1.chr2L 13963658 115 - 22036055 CCUCCAGUGCCAAGGAGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUGUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCCCCAUCGGAAAAAUUCCAAGCCAUAAGGGGGAAAAU- (((((...((((((((((.((--((((((....))))))))..((((.(((....))))))).....))))))))))..........(((.....))).........))))).....- ( -38.60, z-score = -1.39, R) >droGri2.scaffold_15252 4763889 102 - 17193109 ----CAAUUGAAAAGCACUUGCACUUGGCGCUGGCUUGGUAUUUGCAUU---------UGCAUUAACAAAGUGUGACCAAC---AUUGAAAAUGCAUCUCGCAAUGCAAAUGAACAUU ----(((.((..((((...(((.....)))...))))..)).)))((((---------((((((.....(((((.....))---)))......((.....))))))))))))...... ( -23.50, z-score = 0.17, R) >consensus CCUCCAGUGCCAAGGCGACUG__UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCC___AUCGGAAAAAUUCCAAGCCAUAAGGGGGAAAAU_ .....((((((.((....)).....((((....))))))))))....................(((((((((.((((..........(((.....)))..)))).))))))))).... (-17.87 = -18.32 + 0.45)

| Location | 14,220,352 – 14,220,446 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 77.18 |
| Shannon entropy | 0.42184 |
| G+C content | 0.47686 |
| Mean single sequence MFE | -31.63 |
| Consensus MFE | -17.73 |
| Energy contribution | -19.07 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.16 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.56 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.40 |
| SVM RNA-class probability | 0.935947 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
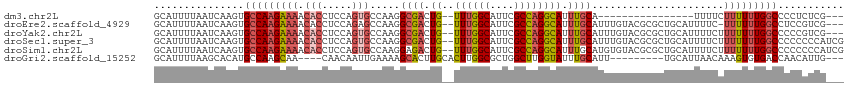
>dm3.chr2L 14220352 94 - 23011544 GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCA----------------UUUUCUUUUUUGGCCCCUCUCG--- ...............(((((((((.(((.....))).....((((.((--((((((....)))))))).)))).----------------.....)))))))))........--- ( -27.10, z-score = -2.28, R) >droEre2.scaffold_4929 15395304 109 + 26641161 GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGAGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUC-UUUUUUGGCCUCCGUCG--- ............((.(((((((((.......(((.((((((((((.((--((((((....)))))))).)))).))))...)).))).......-)))))))))))......--- ( -32.54, z-score = -1.95, R) >droYak2.chr2L 14318792 110 - 22324452 GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCGUCG--- ...............((((((((((......((((((((((((((.((--((((((....)))))))).)))).))))...)))))).......))))))))))........--- ( -36.12, z-score = -3.01, R) >droSec1.super_3 477982 113 - 7220098 GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGUGCCAAGGCGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCCCCAUCG ...............((((((((((......((((((((((((((.((--((((((....)))))))).)))).))))...)))))).......))))))))))........... ( -36.12, z-score = -3.35, R) >droSim1.chr2L 13963687 113 - 22036055 GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGUGCCAAGGAGACUG--UUUGGCAUUCGCCAGGCAUUUGCAUGUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCCCCAUCG ...............((((((((((.......((((((.((....)).--..((((....))))))))))((((.(((....))))))).....))))))))))........... ( -32.90, z-score = -2.05, R) >droGri2.scaffold_15252 4763919 99 - 17193109 GCAUUUUAAGCACAUGCCAAGCAA----CAACAAUUGAAAAGCACUUGCACUUGGCGCUGGCUUGGUAUUUGCAUU---------UGCAUUAACAAAGUGUGACCAACAUUG--- (((......(((.(((((((((..----((.....))...(((.((.......)).))).))))))))).)))...---------)))........(((((.....))))).--- ( -25.00, z-score = -0.06, R) >consensus GCAUUUUAAUCAAGUGCCAAGAAAACACCUCCAGUGCCAAGGCGACUG__UUUGGCAUUCGCCAGGCAUUUGCAUUUGUACGCGCUGCAUUUUCUUUUUUUGGCCCCCCUCG___ ...............(((((((((........((((((.((....)).....((((....))))))))))((((...........))))......)))))))))........... (-17.73 = -19.07 + 1.34)
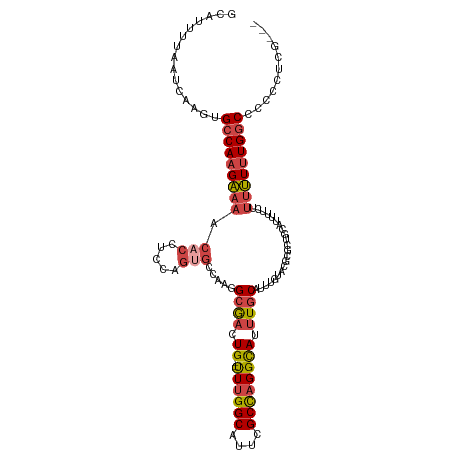
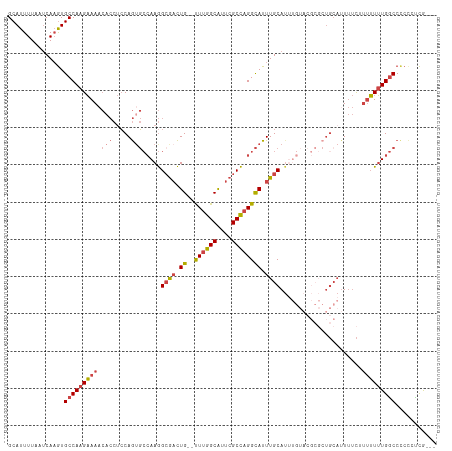
| Location | 14,220,446 – 14,220,499 |
|---|---|
| Length | 53 |
| Sequences | 5 |
| Columns | 54 |
| Reading direction | reverse |
| Mean pairwise identity | 81.94 |
| Shannon entropy | 0.32412 |
| G+C content | 0.55306 |
| Mean single sequence MFE | -17.50 |
| Consensus MFE | -8.90 |
| Energy contribution | -10.06 |
| Covariance contribution | 1.16 |
| Combinations/Pair | 1.27 |
| Mean z-score | -2.30 |
| Structure conservation index | 0.51 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.679241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14220446 53 - 23011544 GCCCAUAGAUUGCAG-UUCCCAAUGGCACUUAUGGCCAUCGAAACGCAGUCGCU .......((((((..-(((...(((((.(....)))))).)))..))))))... ( -16.40, z-score = -2.35, R) >droAna3.scaffold_12916 4613910 52 + 16180835 GCCCCUGAACCUCGG--UUGCAGGCGCACUUGUAGCCACCGCACUGCACAAGCU ((.(((((((....)--)).)))).)).(((((.((.........))))))).. ( -15.40, z-score = -0.38, R) >droEre2.scaffold_4929 15395413 53 + 26641161 GCCCAUAGAUUGCAG-UUCACAGUGGCACUUAUGGCCAUCGAAACGCAGUCGCU .......((((((..-(((...(((((.(....)))))).)))..))))))... ( -16.70, z-score = -2.12, R) >droSec1.super_3 478095 54 - 7220098 GCCCAUAGAUUGCAGUUUCCCAGUGGCACUUAUGGCCAUCGAAACGCAGUCGCU .......((((((.(((((...(((((.(....)))))).)))))))))))... ( -19.50, z-score = -3.32, R) >droSim1.chr2L 13963800 54 - 22036055 GCCCAUAGAUUGCAGUUUCCCAGUGGCACUUAUGGCCAUCGAAACGCAGUCGCU .......((((((.(((((...(((((.(....)))))).)))))))))))... ( -19.50, z-score = -3.32, R) >consensus GCCCAUAGAUUGCAG_UUCCCAGUGGCACUUAUGGCCAUCGAAACGCAGUCGCU .......((((((.(.(((...(((((.......))))).))).)))))))... ( -8.90 = -10.06 + 1.16)

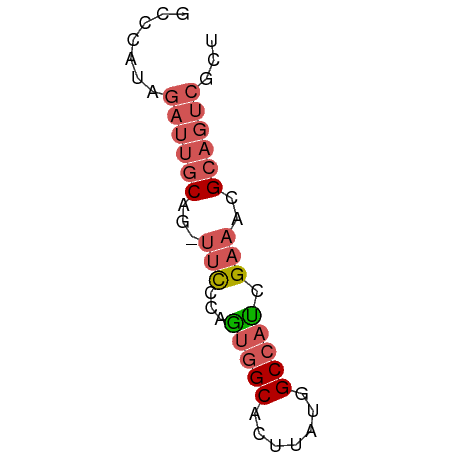
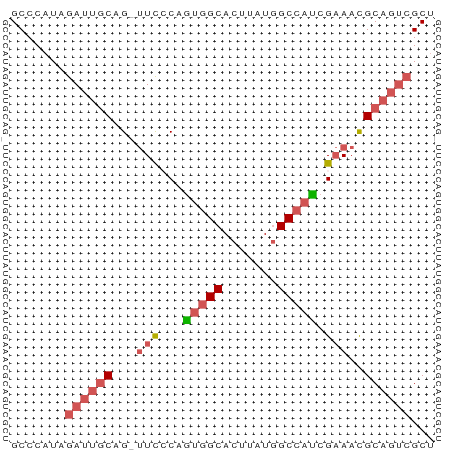
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:40 2011