| Sequence ID | dm3.chr2L |
|---|---|
| Location | 1,293,150 – 1,293,268 |
| Length | 118 |
| Max. P | 0.922046 |

| Location | 1,293,150 – 1,293,268 |
|---|---|
| Length | 118 |
| Sequences | 11 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 75.39 |
| Shannon entropy | 0.55508 |
| G+C content | 0.60280 |
| Mean single sequence MFE | -47.06 |
| Consensus MFE | -18.27 |
| Energy contribution | -18.58 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.52 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.29 |
| SVM RNA-class probability | 0.922046 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 1293150 118 + 23011544 GACUGGGCGCCUAUGCCAACACCACCUUGGCGGCUCAGAUGGCCAGUGGAGGUGGAGCGGGCACCUUGAGGCGCCAGCGUCAGCCGAAGACGCUUUACAAAAGCGAAAACAAUAUACU (((..(((((((.((((..(.(((((((.((((((.....)))).)).))))))).)..)))).....)))))))...))).........(((((.....)))))............. ( -51.90, z-score = -3.76, R) >droSim1.chrU 12843899 118 + 15797150 GACUGGGCGCCUACGCCAACACCACCUUGGCGGCCCAGAUGGCCAGUGGAGGUGGAGCGGGCACCUUGAGGCGCCAGCGGCAGCCCAAAACGCUUUACAAAAGCGAGAACAAUAUACU ...((((((((..........(((((((.((((((.....)))).)).))))))).((.(((.((....)).))).))))).)))))...(((((.....)))))............. ( -53.80, z-score = -3.82, R) >droSec1.super_14 1243547 118 + 2068291 GACUGGGUGCCUACGCCAACACCACCUUGGCGGCCCAGAUGGCCAGUGGAGGUGGAGCGGGCACAUUGAGGCGCCAGCGGCAGCCCAAAACGCUUUACAAAAGCGAGAACAAUAUACU ...(((((((((..(((..(.(((((((.((((((.....)))).)).))))))).)..)))......))))(((...))).)))))...(((((.....)))))............. ( -50.20, z-score = -3.07, R) >droYak2.chr2L 1269140 118 + 22324452 GACUGGGUGCCUAUGCCAACACCACCUUGGCUGCCCAGAUGGCCAGUGGCGGUGGAGCGGGCACCUUGAGGCGCCAGCGGCAGCCCAAGACGCUUUACAAGAGCGAGAACAACAUACU ...((((((((........((((.((((((((........)))))).)).))))..((.(((.((....)).))).)))))).))))...(((((.....)))))............. ( -48.40, z-score = -1.68, R) >droEre2.scaffold_4929 1333051 118 + 26641161 GACUCGGCGCCUACGCCAACACCACCUUGGCGGCCCAGAUGGCCAGUGGCGGCGGAGCGGGCACCUUGAGGCGCCAGCGGCAGCCCAAAACGCUUUACAAAAGCGAGAACAACAUACU ..((((.((((.(((((((.......)))))((((.....)))).))))))(..((((((((.((....)).))).((....))......)))))..).....))))........... ( -45.20, z-score = -1.14, R) >droAna3.scaffold_12916 4755401 118 + 16180835 GUCUGGGCGCCUAUGCCAAUACCACACUGGUGGCCCAAAUGGCGAACGGCGGCGGGGCCGGGACUCUGAGACGGCAACGGCAGCCGAAAACGCUCUACAAAAGCGAAAAUAACAUUUU ((((.(((.((..((((..((((.....))))(((.....)))....))))..)).))).)))).......((((.......))))....((((.......))))............. ( -41.30, z-score = -1.21, R) >droWil1.scaffold_180708 2922210 118 + 12563649 GUCUUGGCGCCUAUGCCAAUACAACAUUGGCCGCCCAGAUGGCCAACGGUGGUGGCGUUGGUACACUUCGUCGUCAGCGUCAACCGAAGGCUCUCUAUAAGAGUGAAAACAAUAUUCU .((((((((.....((((((.....)))))))))).(((.((((..((((..((((((((((((.....)).))))))))))))))..)))).)))..))))((....))........ ( -39.40, z-score = -1.88, R) >droMoj3.scaffold_6500 28675141 118 - 32352404 GCCUGGGCGCCUAUGCGAACACCACGCUGGCGGCUCAAAUAGCCAGCGGCGGUGGGGCCGGCACGCUGAGGCGCCAGCGUCAGCCGAAGACGCUCUUCAAGAGCGAGAACAACAUCCU ((((...(((....)))..((((.(((((((..........)))))))..))))))))((((((((((......))))))..))))....(((((.....)))))............. ( -52.60, z-score = -1.46, R) >droVir3.scaffold_12963 7229115 118 + 20206255 GCCUCGGCGCCUAUGCAAACACCACGCUGGCCGCCCAAAUGGCCAGCGGCGGCGGAGCUGGCACCCUGCGCCGGCAGCGCCAGCCGAAGACGCUCUUCAAGAGCGAGACCAACAUAUU ...((((((((..((....))....((((((((......)))))))))))((((..((((((.......))))))..)))).)))))...(((((.....)))))............. ( -55.70, z-score = -3.08, R) >droGri2.scaffold_15252 10369818 118 + 17193109 GCUUGGGCGCCUAUGCGAAUACCACACUGGCGGCUCAAAUGGCCAGCGGUGGUGGGGCGGGCACCUUGCGCCGCCAACGACAGCCUAAAACGCUCUUCAAGAGCGAGAACAACAUAUU ..((((((((....))...((((((.(((((.(......).)))))..)))))).(((((((.....)).))))).......))))))..(((((.....)))))............. ( -46.20, z-score = -1.66, R) >anoGam1.chr3R 1126392 98 - 53272125 ------GUUCAUCGUCGGGCCCUAC---GGCAAGCUAUAU---CACCGGUGGUGGCAUGGGCUCGUCGUA-CGGUGGC-UCAACGGUAAGUAAUCU-CAACAGUGGAAGCCAC----- ------....(((((.(((((((((---(((.((((((..---((((...))))..)).)))).))))).-.)).)))-)).))))).........-.....((((...))))----- ( -33.00, z-score = -1.12, R) >consensus GACUGGGCGCCUAUGCCAACACCACCUUGGCGGCCCAGAUGGCCAGCGGCGGUGGAGCGGGCACCUUGAGGCGCCAGCGGCAGCCGAAAACGCUCUACAAAAGCGAGAACAACAUACU .....((((((...((...(.((.(.(((((..........))))).)..)).)..)).))).......(.((....)).).))).....(((((.....)))))............. (-18.27 = -18.58 + 0.31)

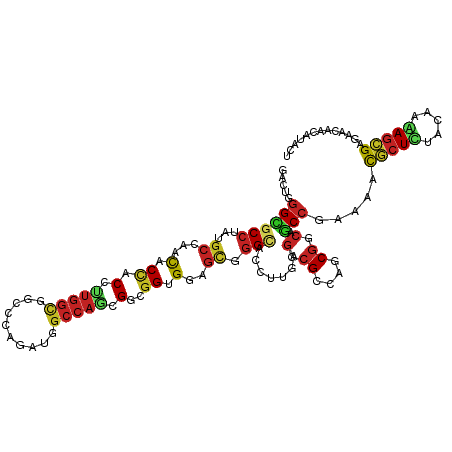

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:08:13 2011