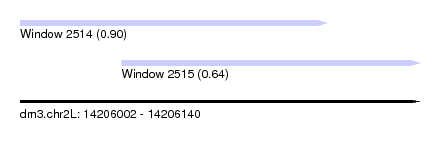
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,206,002 – 14,206,140 |
| Length | 138 |
| Max. P | 0.895105 |

| Location | 14,206,002 – 14,206,108 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 63.09 |
| Shannon entropy | 0.79479 |
| G+C content | 0.46311 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -10.23 |
| Energy contribution | -10.40 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.76 |
| Mean z-score | -1.04 |
| Structure conservation index | 0.41 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.12 |
| SVM RNA-class probability | 0.895105 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
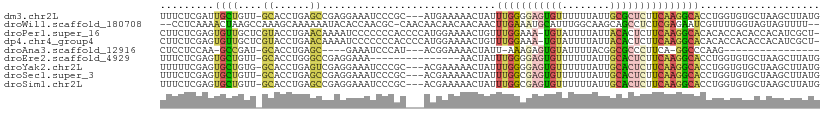
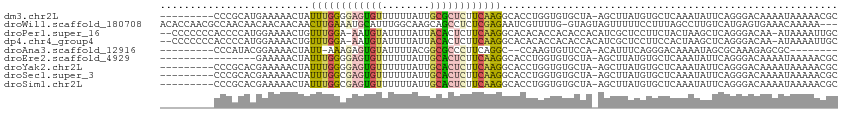
>dm3.chr2L 14206002 106 + 23011544 UUUCUCGAUUGCUGUU-GCACCUGAGCCGAGGAAAUCCCGC---AUGAAAAACUAUUUGGGGAGUGUUUUUUAUUGCGCUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG ..........(((..(-(((((.(.((((.((....)))..---............(((((((((((........)))))))))))))).)..))))))...)))..... ( -31.20, z-score = -1.36, R) >droWil1.scaffold_180708 1263894 105 + 12563649 --CCUCAAAACUAAGCCAAAGCAAAAAAUACACCAACGC-CAACAACAACAACAACUUGAAAUGCAUUUGGCAAGCAGCCUCUCGAGAAUCGUUUUGGUAGUAGUUUU-- --....(((((((.((((((((...............((-(((...((.(((....)))...))...))))).......((....))....))))))))..)))))))-- ( -19.60, z-score = -1.29, R) >droPer1.super_16 1533472 108 + 1990440 CUUCUCGAGUGUUGCUCGUACCUGAACAAAAUCCCCCCCACCCCAUGGAAAACUGUUUGGAAA-UGUAUUUUAUUACACUCUUCAAGGCACACACCACACCACAUCGCU- ......(.((((((((.....(..((((...(((............)))....))))..)...-((((......))))........)))).))))).............- ( -15.40, z-score = 0.40, R) >dp4.chr4_group4 1168059 108 - 6586962 CUUCUCGAGUGUUGCUCGUACCUGAACAAAAUCCCCCCCACCCCAUGGAAAACUGUUUGGAAA-UGUAUUUUAUUACACUCUUCAAGGCACACACCACACCACAUCGCU- ......(.((((((((.....(..((((...(((............)))....))))..)...-((((......))))........)))).))))).............- ( -15.40, z-score = 0.40, R) >droAna3.scaffold_12916 4601183 83 - 16180835 CUCCUCCAA-GCCGAU-GCACCUGAGC----GAAAUCCCAU---ACGGAAAACUAUU-AAAGAGUGUAUUUUACGGCGCCCUUCA-GGCCCAAG---------------- .........-((((((-((((((....----....(((...---..)))........-..)).))))))....))))(((.....-))).....---------------- ( -17.17, z-score = -0.82, R) >droEre2.scaffold_4929 15379657 94 - 26641161 UUUCUCGAGUGCUGUU-GCACCUGGGCCGAGGAAA---------------AACUAUUUGGGGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG ......((((...((.-(((((.(((((.((....---------------..))..(((((((((((........)))))))))))))).)).)))))))...))))... ( -34.20, z-score = -3.02, R) >droYak2.chr2L 14304227 106 + 22324452 UUUUUCGAGUGCUGUG-GCACCUGAGUCGAGGAAAUCCCGC---ACGAAAAACUAUUUGGGGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG (((((((.((((....-))))....(.((.((....)))))---.)))))))....(((((((((((........)))))))))))(((((.....)))))......... ( -33.10, z-score = -1.73, R) >droSec1.super_3 463578 106 + 7220098 UUUCUCGAGUGCUGUU-GCACCUGAGCCGAGGAAAUCCCGC---ACGAAAAACUAUUUGGCGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG ........((((....-)))).(((((.(.((....)))((---(((........(((((.((((((........)))))).)))))........)))))...))))).. ( -29.69, z-score = -0.99, R) >droSim1.chr2L 13949307 106 + 22036055 UUUCUCGAGUGCUGUU-GCACCUGAGCCGAGGAAAUCCCGC---ACGAAAAACUAUUUGGCGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG ........((((....-)))).(((((.(.((....)))((---(((........(((((.((((((........)))))).)))))........)))))...))))).. ( -29.69, z-score = -0.99, R) >consensus UUUCUCGAGUGCUGUU_GCACCUGAGCCGAGGAAAUCCCGC___ACGAAAAACUAUUUGGAGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUAAGCUUAUG .........((((....((......)).............................(((((((((((........))))))))))))))).................... (-10.23 = -10.40 + 0.17)
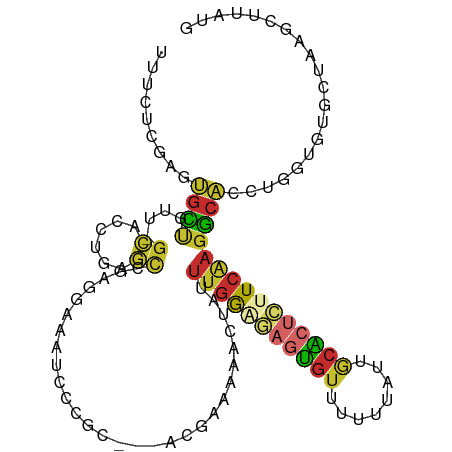
| Location | 14,206,037 – 14,206,140 |
|---|---|
| Length | 103 |
| Sequences | 9 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 63.34 |
| Shannon entropy | 0.75489 |
| G+C content | 0.42607 |
| Mean single sequence MFE | -21.65 |
| Consensus MFE | -10.00 |
| Energy contribution | -10.27 |
| Covariance contribution | 0.26 |
| Combinations/Pair | 1.75 |
| Mean z-score | -0.50 |
| Structure conservation index | 0.46 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.635792 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14206037 103 + 23011544 ---------CCCGCAUGAAAAACUAUUUGGGGAGUGUUUUUUAUUGCGCUCUUCAAGGCACCUGGUGUGCUA-AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC ---------((((((((........((((((((((((........))))))))))))........)))))..-(((......)))..........)))............... ( -26.69, z-score = -1.17, R) >droWil1.scaffold_180708 1263921 109 + 12563649 ACACCAACGCCAACAACAACAACAACUUGAAAUGCAUUUGGCAAGCAGCCUCUCGAGAAUCGUUUUG-GUAGUAGUUUUUCCUUUAGCCUUGUCAUGAGUGAAACAAAAA--- .(((((..(((((...((.(((....)))...))...)))))..((.(((...((.....))....)-)).))......................)).))).........--- ( -14.10, z-score = 2.31, R) >droPer1.super_16 1533504 109 + 1990440 --CCCCCCCACCCCAUGGAAAACUGUUUGGA-AAUGUAUUUUAUUACACUCUUCAAGGCACACACCACACCACAUCGCUCCUUCUACUAAGCUCAGGGACAA-AUAAAAUUGC --.........(((.(((........(((((-..((((......))))...)))))((......))...)))....(((..........)))...)))....-.......... ( -13.50, z-score = 1.19, R) >dp4.chr4_group4 1168091 109 - 6586962 --CCCCCCCACCCCAUGGAAAACUGUUUGGA-AAUGUAUUUUAUUACACUCUUCAAGGCACACACCACACCACAUCGCUCCUUCCACUAAGCUCAGGGACAA-AUAAAAUUGC --.........(((.(((........(((((-..((((......))))...)))))((......))...)))....(((..........)))...)))....-.......... ( -13.50, z-score = 1.29, R) >droAna3.scaffold_12916 4601213 92 - 16180835 ---------CCCAUACGGAAAACUAUU-AAAGAGUGUAUUUUACGGCGCCCUUCAGGC--CCAAGUGUUCCA-ACAUUUCAGGGACAAAAUAGCGCAAAGAGCGC-------- ---------(((....((((.(((...-.....(((.....)))((.(((.....)))--)).))).)))).-........)))........((((.....))))-------- ( -20.32, z-score = -0.68, R) >droEre2.scaffold_4929 15379687 96 - 26641161 ----------------GAAAAACUAUUUGGGGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUA-AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC ----------------.........((((((((((((........))))))))))))(..((((((((....-(((......)))...))).)))))..)............. ( -25.40, z-score = -2.26, R) >droYak2.chr2L 14304262 103 + 22324452 ---------CCCGCACGAAAAACUAUUUGGGGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUA-AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC ---------((((((((........((((((((((((........))))))))))))........)))))..-(((......)))..........)))............... ( -29.59, z-score = -2.43, R) >droSec1.super_3 463613 103 + 7220098 ---------CCCGCACGAAAAACUAUUUGGCGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUA-AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC ---------((((((((........(((((.((((((........)))))).)))))........)))))..-(((......)))..........)))............... ( -25.89, z-score = -1.37, R) >droSim1.chr2L 13949342 103 + 22036055 ---------CCCGCACGAAAAACUAUUUGGCGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUA-AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC ---------((((((((........(((((.((((((........)))))).)))))........)))))..-(((......)))..........)))............... ( -25.89, z-score = -1.37, R) >consensus _________CCCGCACGAAAAACUAUUUGGAGAGUGUUUUUUAUUGCACUCUUCAAGGCACCUGGUGUGCUA_AGCUUAUGUGCUCAAAUAUUCAGGGACAAAAUAAAAACGC .........................((((((((((((........))))))))))))........................................................ (-10.00 = -10.27 + 0.26)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:37 2011