| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,177,380 – 14,177,486 |
| Length | 106 |
| Max. P | 0.634756 |

| Location | 14,177,380 – 14,177,486 |
|---|---|
| Length | 106 |
| Sequences | 9 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 71.40 |
| Shannon entropy | 0.59790 |
| G+C content | 0.47397 |
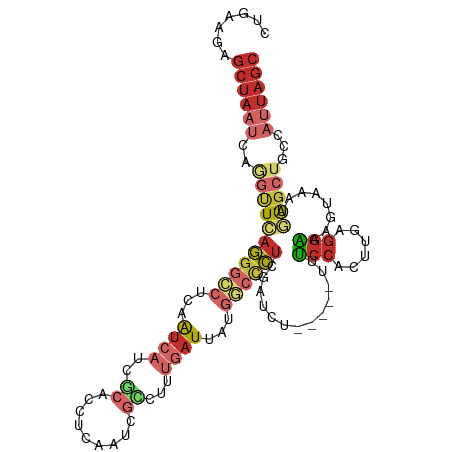
| Mean single sequence MFE | -27.90 |
| Consensus MFE | -13.39 |
| Energy contribution | -15.20 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.02 |
| Structure conservation index | 0.48 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.634756 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
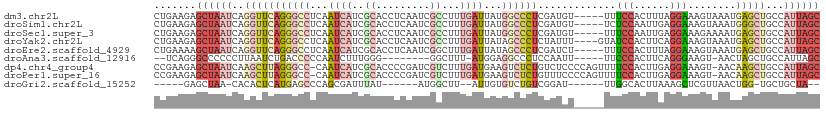
>dm3.chr2L 14177380 106 - 23011544 CUGAAGAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGCCUUUGAUUAUGGCCCUCGAUGU-----UUUCCACUUUAGGAAAGUAAAUGAGCUGCCAUUAGC .......((((((..(((((((((((..(((((..((.........))...)))))..)))))......(-----(((((......))))))....))))))...)))))) ( -32.80, z-score = -2.45, R) >droSim1.chr2L 13920555 106 - 22036055 CUGAAGAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGCCUUUGAUUAUGGCCCUCGAUGU-----UCUCCAAUUGAGGAAAGUAAAUGGGCUGCCAUUAGC .......((((((..(((((((((((..(((((..((.........))...)))))..)))))......(-----((((.....))))).......))))))...)))))) ( -30.30, z-score = -0.79, R) >droSec1.super_3 435021 106 - 7220098 CUGAAGAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGCCUUUGAUUAUGGCCCUCGAUGU-----UUUCCAAUUGAGGAAAGAAAAUGAGCUGCCAUUAGC .......((((((..(((((((((((..(((((..((.........))...)))))..)))))......(-----(((((......))))))....))))))...)))))) ( -33.00, z-score = -2.08, R) >droYak2.chr2L 14275383 107 - 22324452 CUGAAGAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGCCUUUGAUUAUAGCCCUCUAUUU----GUAUCCACUUCAGGAAAGUAAAUGAGCUGCCAUUAGC .......((((((..((((((((((...(((((..((.........))...)))))...)))).......----...(((......))).......))))))...)))))) ( -25.90, z-score = -0.92, R) >droEre2.scaffold_4929 15351144 106 + 26641161 CUGAAAAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGGCUUUGAUUAUAGCCCUCGAUCU-----UUUCCACUUUAGGAAAGUAAAUGAGCUGCCAUUAGC .......((((((..((((((((((...(((((..((..........))..)))))...))))......(-----(((((......))))))....))))))...)))))) ( -29.00, z-score = -1.92, R) >droAna3.scaffold_12916 4572764 94 + 16180835 --UCAGGGCCCCCCUUAAUCUGACCCCCAAUCUUUGGG--------GGCUUU-AUGGAGGCCCUCCAAUU-----UUCCCACUUCAGGGAAGU-AACUAGCUGCCAUUAGC --..((((((.((..(((...(.((((((.....))))--------))).))-).)).)))))).....(-----(((((......)))))).-.....((((....)))) ( -34.20, z-score = -1.51, R) >dp4.chr4_group4 1130612 109 + 6586962 CCGAAGAGCUAAUCAAGCUUAGGGCC-CAAUCAUCGCACCCCGAUCGUCUUUGAUGAAGUCUCUGUCUCCCCAGUUUUCCACUUGAGGAAAGU-AACAAGCUGCCAUUAGC .......((((((..(((((.(((..-......(((.....)))(((((...)))))............)))..((((((......)))))).-...)))))...)))))) ( -22.70, z-score = 0.15, R) >droPer1.super_16 1496910 109 - 1990440 CCGAAGAGCUAAUCAAGCUUAGGGCC-CAAUCAUCGCACCCCGAUCGUCUUUGAUGAAGUCUCUGUUUCCCCAGUUUUCCACUUGAGGAAAGU-AACAAGCUGCCAUUAGC .......((((((..(((((.(((..-......(((.....)))(((((...)))))............)))..((((((......)))))).-...)))))...)))))) ( -22.70, z-score = 0.01, R) >droGri2.scaffold_15252 12481788 88 + 17193109 -----GAGCUAA-CACACUCAUGAGCCCAGCGAUUUAU------AUGGCUU--AUUGUGUCUGUCGGAU------UUGGCACUUAAAGCUCGUUAACUGG-UGCUGCUA-- -----(((....-....)))...(((((((....(((.------(((((((--...(((((........------..)))))...)))).))))))))))-.)))....-- ( -20.50, z-score = 0.35, R) >consensus CUGAAGAGCUAAUCAGGUUCAGGGCCUCAAUCAUCGCACCUCAAUCGCCUUUGAUUAUGGCCCUCGAUCU_____UUUCCACUUGAGGAAAGUAAAUGAGCUGCCAUUAGC .......((((((..(((((((((((...((((..((.........))...))))...)))))).............(((......)))........)))))...)))))) (-13.39 = -15.20 + 1.81)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:33 2011