
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,160,905 – 14,161,008 |
| Length | 103 |
| Max. P | 0.815879 |

| Location | 14,160,905 – 14,161,008 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.39381 |
| G+C content | 0.26697 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -9.14 |
| Energy contribution | -10.20 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.06 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.78 |
| SVM RNA-class probability | 0.815879 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
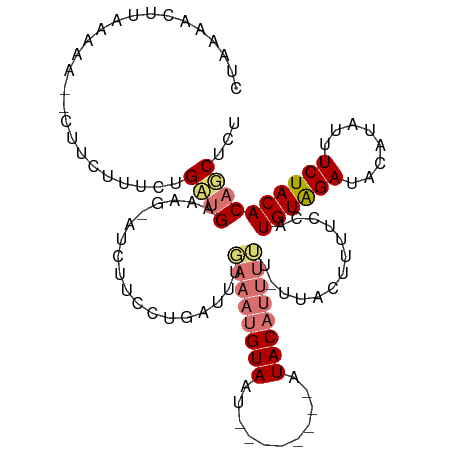
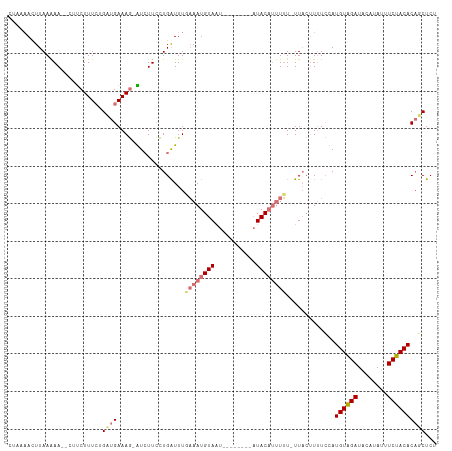
>dm3.chr2L 14160905 103 + 23011544 CGUUAAUUUAAAAA--UGCCUUUGUGAUGAAAG-GUCUUCCUGAUUUUAAAUGUAAU--------AUACAUUUUUCUUACAUAUCCAUGUAGAUACAUAUGUCUACACAACUCU .(((.........(--((....(((((.(((((-(....)))......(((((((..--------.)))))))))))))))....)))(((((((....)))))))..)))... ( -16.50, z-score = -1.37, R) >droEre2.scaffold_4929 15332703 101 - 26641161 -UUUUAUUUAAGAAUACUUCUUUCUGAUGAAAAUGUGUUUUUGGUUUUUAAUGUA-----------UACCACUGU-UUUGUUUUACAUGUGGAUUCAGAUUUCUACACCUCUCU -........((((((((...((((....))))..))))))))(((.....(((((-----------....((...-...))..)))))(((((........))))))))..... ( -16.20, z-score = -0.98, R) >droSec1.super_3 419129 110 + 7220098 CUAAAACUUAAAAA--CUUCUUUCUGCUGAAAG-AUCUUCCUGAUUUGAAAUGUAAUUUGUUAUUAUACAUUUUU-UUACUUUUCUAUGUAGAUACAUAUUUCUACACAGCUCU ..............--.........((((..((-(((.....)))))((((.((((..(((......))).....-)))).))))..((((((........))))))))))... ( -18.30, z-score = -2.94, R) >droSim1.chr2L_random 553084 110 + 909653 CUAAAACUUAAAAA--CUUCUUUCUGCUGAAAG-AUCUUCCUGAUUUGAAAUGUAAUUUGUUAUUAUACAUUUUU-UUACUUUUCUAUGUAGAUACAUAUUUCUACACAGCUCU ..............--.........((((..((-(((.....)))))((((.((((..(((......))).....-)))).))))..((((((........))))))))))... ( -18.30, z-score = -2.94, R) >consensus CUAAAACUUAAAAA__CUUCUUUCUGAUGAAAG_AUCUUCCUGAUUUGAAAUGUAAU________AUACAUUUUU_UUACUUUUCCAUGUAGAUACAUAUUUCUACACAGCUCU .........................((((..................((((((((...........)))))))).............((((((........))))))))))... ( -9.14 = -10.20 + 1.06)

| Location | 14,160,905 – 14,161,008 |
|---|---|
| Length | 103 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 74.40 |
| Shannon entropy | 0.39381 |
| G+C content | 0.26697 |
| Mean single sequence MFE | -17.32 |
| Consensus MFE | -10.62 |
| Energy contribution | -11.12 |
| Covariance contribution | 0.50 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.61 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.07 |
| SVM RNA-class probability | 0.527420 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
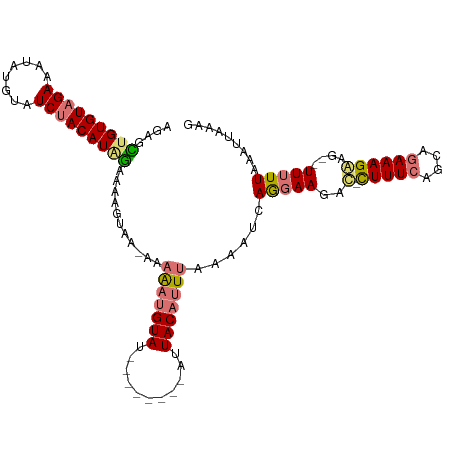
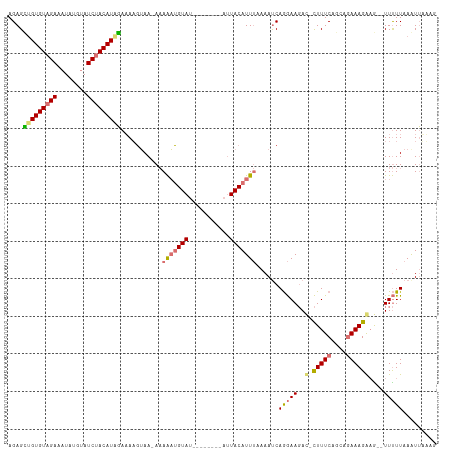
>dm3.chr2L 14160905 103 - 23011544 AGAGUUGUGUAGACAUAUGUAUCUACAUGGAUAUGUAAGAAAAAUGUAU--------AUUACAUUUAAAAUCAGGAAGAC-CUUUCAUCACAAAGGCA--UUUUUAAAUUAACG .((.(..((((((........))))))..)...........(((((((.--------..)))))))....))(((((..(-((((......)))))..--)))))......... ( -15.80, z-score = -0.37, R) >droEre2.scaffold_4929 15332703 101 + 26641161 AGAGAGGUGUAGAAAUCUGAAUCCACAUGUAAAACAAA-ACAGUGGUA-----------UACAUUAAAAACCAAAAACACAUUUUCAUCAGAAAGAAGUAUUCUUAAAUAAAA- .(((((((((((....))....((((.(((........-)))))))..-----------.................)))).(((((....))))).....)))))........- ( -13.10, z-score = -0.70, R) >droSec1.super_3 419129 110 - 7220098 AGAGCUGUGUAGAAAUAUGUAUCUACAUAGAAAAGUAA-AAAAAUGUAUAAUAACAAAUUACAUUUCAAAUCAGGAAGAU-CUUUCAGCAGAAAGAAG--UUUUUAAGUUUUAG ....(((((((((........))))))))).....(((-(((((((((...........)))))))......(((((..(-(((((....))))))..--)))))...))))). ( -20.20, z-score = -2.27, R) >droSim1.chr2L_random 553084 110 - 909653 AGAGCUGUGUAGAAAUAUGUAUCUACAUAGAAAAGUAA-AAAAAUGUAUAAUAACAAAUUACAUUUCAAAUCAGGAAGAU-CUUUCAGCAGAAAGAAG--UUUUUAAGUUUUAG ....(((((((((........))))))))).....(((-(((((((((...........)))))))......(((((..(-(((((....))))))..--)))))...))))). ( -20.20, z-score = -2.27, R) >consensus AGAGCUGUGUAGAAAUAUGUAUCUACAUAGAAAAGUAA_AAAAAUGUAU________AUUACAUUUAAAAUCAGGAAGAC_CUUUCAGCAGAAAGAAG__UUUUUAAAUUAAAG ....(((((((((........)))))))))...........(((((((...........)))))))...............(((((....)))))................... (-10.62 = -11.12 + 0.50)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:32 2011