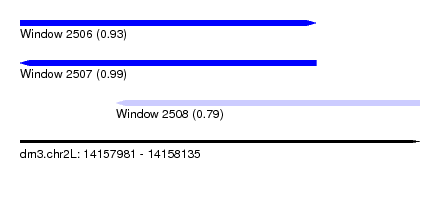
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,157,981 – 14,158,135 |
| Length | 154 |
| Max. P | 0.993102 |

| Location | 14,157,981 – 14,158,095 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 58.28 |
| Shannon entropy | 0.68370 |
| G+C content | 0.67302 |
| Mean single sequence MFE | -52.58 |
| Consensus MFE | -24.94 |
| Energy contribution | -29.69 |
| Covariance contribution | 4.75 |
| Combinations/Pair | 1.38 |
| Mean z-score | -1.21 |
| Structure conservation index | 0.47 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.34 |
| SVM RNA-class probability | 0.928190 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
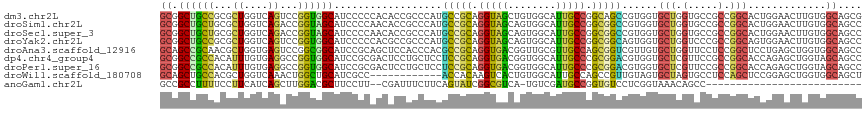
>dm3.chr2L 14157981 114 + 23011544 CCGCUGCUGCAGCCGCGGCUGCGGCCGCAGCGGUGGUCGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACGGCUGCCGGCAAUGCCACAGCUACCUGCGGCAUGGGCG ((((((((((.(((((....))))).)))))))))).....(((...........((((((((((............))))))))))(((((.(((....))).))))).))). ( -65.50, z-score = -1.70, R) >droYak2.chr2L 14255795 114 + 22324452 CUGCUGCAGCAGCCGCGGCUGCGGCCGCAGCUGUAGUGGCUGCCACAAGUUCCACUGCCGGCGGGACCAGCACCACUGCCGCCGGCAAUGCCACUGCUACCUGCGGCAUGGGCG ..(((...(((((....))))).(((((((..((((((((.......((.....))((((((((...(((.....)))))))))))...))))))))...)))))))...))). ( -62.00, z-score = -1.64, R) >droWil1.scaffold_180708 11236551 106 - 12563649 CCGCGGCAGCAGCUGCUGCGGCUGCUGCAGCAGUUGCAGCUGCCACCAGCUCCGGAGCUGGAGGCACUAGCACUACAACGGCUGGCAAUGCCACAGUGACUUGUGG-------- (((((((.(((((((((((((...))))))))))))).)))))..((((((....)))))).))(((.((((((.....(((.......)))..)))).)).))).-------- ( -54.30, z-score = -1.49, R) >anoGam1.chr2L 23305266 96 + 48795086 ---GGGCAGGAGAUCGAGCCGAAGGUGGCGGAGGAGCUG-UGUCGGCUGUU----UACCGAGGACACCGGCAUCGACAUGAC-GCCGAUACUGAAGAAAUCGAAG--------- ---...(((...(((..(((...)))((((..(...(.(-((((((.((((----(.....)))))))))))).)...)..)-)))))).)))............--------- ( -28.50, z-score = -0.01, R) >consensus CCGCGGCAGCAGCCGCGGCUGCGGCCGCAGCAGUAGCAGCUGCCACAAGUUCCAGUGCCGGCGGCACCAGCACCACAACGGCCGGCAAUGCCACAGCUACCUGCGG________ ((((((((((.(((((((((((....))))))))))).))))))............((((((((..............))))))))................))))........ (-24.94 = -29.69 + 4.75)

| Location | 14,157,981 – 14,158,095 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 58.28 |
| Shannon entropy | 0.68370 |
| G+C content | 0.67302 |
| Mean single sequence MFE | -53.88 |
| Consensus MFE | -29.34 |
| Energy contribution | -30.77 |
| Covariance contribution | 1.44 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.57 |
| Structure conservation index | 0.54 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.59 |
| SVM RNA-class probability | 0.993102 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>dm3.chr2L 14157981 114 - 23011544 CGCCCAUGCCGCAGGUAGCUGUGGCAUUGCCGGCAGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCGACCACCGCUGCGGCCGCAGCCGCGGCUGCAGCAGCGG (((...((((((((((.((((((((...))).)))))...((((((((.....))))))))...))))))))))))).((.(.(((((((((((....))))))))))).).)) ( -68.80, z-score = -1.83, R) >droYak2.chr2L 14255795 114 - 22324452 CGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCAGUGGUGCUGGUCCCGCCGGCAGUGGAACUUGUGGCAGCCACUACAGCUGCGGCCGCAGCCGCGGCUGCUGCAGCAG ......((((((((((.((....))((((((((((((((.....)))...)))))))))))...))))))))))((..(..((((((((((....))))))))))..)..)).. ( -64.00, z-score = -1.22, R) >droWil1.scaffold_180708 11236551 106 + 12563649 --------CCACAAGUCACUGUGGCAUUGCCAGCCGUUGUAGUGCUAGUGCCUCCAGCUCCGGAGCUGGUGGCAGCUGCAACUGCUGCAGCAGCCGCAGCAGCUGCUGCCGCGG --------..........((((((((.(((.(((.((((((((.....((((.((((((....)))))).)))))))))))).))))))(((((.......))))))))))))) ( -55.20, z-score = -1.80, R) >anoGam1.chr2L 23305266 96 - 48795086 ---------CUUCGAUUUCUUCAGUAUCGGC-GUCAUGUCGAUGCCGGUGUCCUCGGUA----AACAGCCGACA-CAGCUCCUCCGCCACCUUCGGCUCGAUCUCCUGCCC--- ---------..((((.......(((..((((-(((.....)))))))(((((...(((.----....)))))))-).))).....(((......)))))))..........--- ( -27.50, z-score = -1.46, R) >consensus ________CCGCAGGUAGCUGUGGCAUUGCCGGCCGCAGUGGUGCUGGUGCCGCCGGCACUGGAACUGGUGGCAGCAACUACUGCUGCAGCCGCAGCCGCAGCUGCUGCAGCGG .....................(((((((((........))))))))).((((((((((......)))))))))).........(((((((((((....)))))))))))..... (-29.34 = -30.77 + 1.44)


| Location | 14,158,018 – 14,158,135 |
|---|---|
| Length | 117 |
| Sequences | 9 |
| Columns | 117 |
| Reading direction | reverse |
| Mean pairwise identity | 68.10 |
| Shannon entropy | 0.70576 |
| G+C content | 0.67925 |
| Mean single sequence MFE | -55.05 |
| Consensus MFE | -21.20 |
| Energy contribution | -20.12 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.79 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.39 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.71 |
| SVM RNA-class probability | 0.793498 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
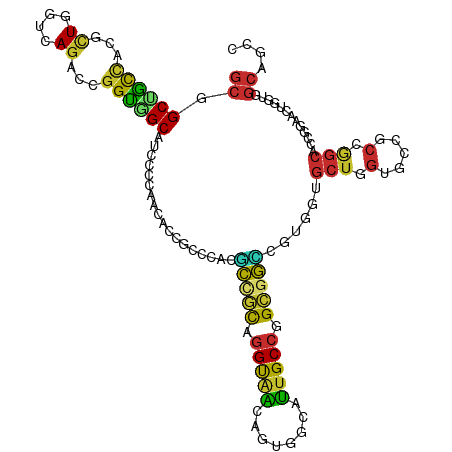
>dm3.chr2L 14158018 117 - 23011544 GCGGCUGCCGCGCUGGUCAGUCCGGUGGCAUCCCCCACACCGCCCAUGCCGCAGGUAGCUGUGGCAUUGCCGGCAGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCG ...(((((((((....((((((((((((((((.(((((.(.(((.(((((((((....)))))))))....))).)..)))).)..))))))))))).)))))....))))))))). ( -69.10, z-score = -2.61, R) >droSim1.chr2L 13908047 117 - 22036055 GCGGCUGCUGCGCUGGUCAGACCGGUAGCAUCCCCAACACCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCC ..((((((..((....((((.((((..(((...(((.((((((....(((((.((((((....))..)))).))))).)))))).))))))..))))..))))....))..)))))) ( -61.40, z-score = -1.09, R) >droSec1.super_3 416151 117 - 7220098 GCGGCUGCUGCGCUGGUCAGACCGGUAGCAUCCCCAACACCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACUGGAACUUGUGGCAGCC ..((((((..((....((((.((((..(((...(((.((((((....(((((.((((((....))..)))).))))).)))))).))))))..))))..))))....))..)))))) ( -61.40, z-score = -1.09, R) >droYak2.chr2L 14255832 117 - 22324452 GCGGCUGCCGCGCUGGUCAGUCCGGUGGCAUCCCCCACGCCGCCCAUGCCGCAGGUAGCAGUGGCAUUGCCGGCGGCAGUGGUGCUGGUCCCGCCGGCAGUGGAACUUGUGGCAGCC ..((((((((((....(((.((((((((.(((.(((((((((((.(((((((........)))))))....)))))).)))).)..))).))))))).).)))....)))))))))) ( -66.20, z-score = -1.87, R) >droAna3.scaffold_12916 4555451 117 + 16180835 GCAGCCGCAACGCUGGUGAGUCCGGCGGCAUCCGCAGCUCCACCCACGCCGCAGGUGACGGUUGCGUUGCCAGCGGUCGUUGUGCUGGUUCCUCCGGCUCCUGAGCUGGUGGCAGCC ((.(((((..((((((.....)))))).......((((((.....(((((((.((..(((....)))..)).)))).)))...(((((.....)))))....))))))))))).)). ( -57.90, z-score = -1.16, R) >dp4.chr4_group4 1104179 117 + 6586962 GCGGCCGCCACAUUUGUGAGGCCGGUGGCAUCCGCGACUCCUGCUCCUCCGCAGGUGACGGUGGCAUUGCCCGCGGACGUGGUGCUCGUUCCGCCGGCACCAGAGCUGGUAGCAGCC .(((((..(((....))).)))))((.((....)).))..(((((((..(...((((.((((((.((.((((((....)))).))..)).)))))).))))...)..)).))))).. ( -53.00, z-score = 0.02, R) >droPer1.super_16 1465868 117 - 1990440 GCGGCCGCCACAUUUGUGAGGCCGGUGGCAUCCGCGACUCCUGCUCCUCCGCAGGUGACGGUGGCAUUGCCCGCGGACGUGGUGCUCGUUCCGCCGGCACCAGAGCUGGUAGCAGCC .(((((..(((....))).)))))((.((....)).))..(((((((..(...((((.((((((.((.((((((....)))).))..)).)))))).))))...)..)).))))).. ( -53.00, z-score = 0.02, R) >droWil1.scaffold_180708 11236588 105 + 12563649 GCAGCUGCCACGCUGGUCAAACUGGCUGCAUCGCC------------ACCACAAGUCACUGUGGCAUUGCCAGCCGUUGUAGUGCUAGUGCCUCCAGCUCCGGAGCUGGUGGCAGCU ..(((((((((((((((..(((.(((((((..(((------------((...........)))))..)).)))))))).....)))))))...((((((....)))))))))))))) ( -49.90, z-score = -2.05, R) >anoGam1.chr2L 23305306 88 - 48795086 GCCGCCUUUUCCUUCAUCAGCUUGGACGCUUCCUU--CGAUUUCUUCAGUAUCGGCGUCA-UGUCGAUGCCGGUGUCCUCGGUAAACAGCC-------------------------- (((((..............))..((((((......--.((.....)).....(((((((.-....)))))))))))))..)))........-------------------------- ( -23.54, z-score = -1.30, R) >consensus GCGGCUGCCACGCUGGUCAGACCGGUGGCAUCCCCAACACCGCCCACGCCGCAGGUAACAGUGGCAUUGCCGGCGGCCGUGGUGCUGGUGCCGCCGGCACCGGAACUGGUGGCAGCC ((.((((((...((....))...))))))..................(((((.(((((........))))).)))))......(((.(.....).))).............)).... (-21.20 = -20.12 + -1.08)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:31 2011