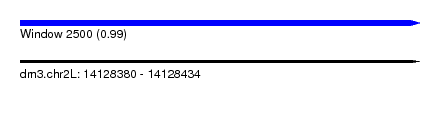
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,128,380 – 14,128,434 |
| Length | 54 |
| Max. P | 0.991638 |

| Location | 14,128,380 – 14,128,434 |
|---|---|
| Length | 54 |
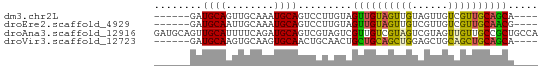
| Sequences | 4 |
| Columns | 64 |
| Reading direction | forward |
| Mean pairwise identity | 62.99 |
| Shannon entropy | 0.55894 |
| G+C content | 0.49074 |
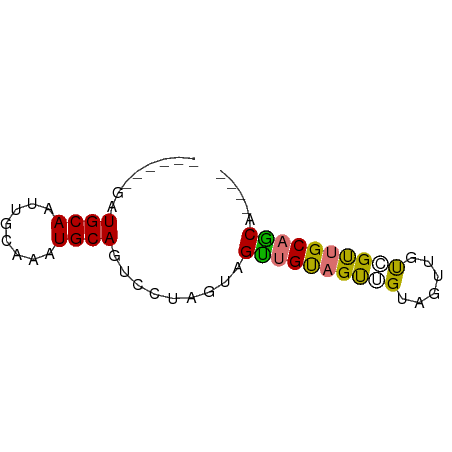
| Mean single sequence MFE | -19.32 |
| Consensus MFE | -11.35 |
| Energy contribution | -11.10 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.50 |
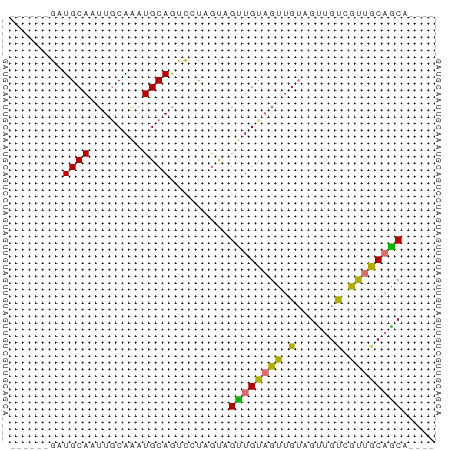
| Mean z-score | -1.92 |
| Structure conservation index | 0.59 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 2.49 |
| SVM RNA-class probability | 0.991638 |
| Prediction | RNA |
| WARNING | Out of training range. z-scores are NOT reliable. |
Download alignment: ClustalW | MAF
>dm3.chr2L 14128380 54 + 23011544 ------GAUGCAGUUGCAAAUGCAGUCCUUGUAGUUGUAGUUGUAGUUGUCGUUGCAGCA---- ------..((((..(((((.(((((...))))).)))))..))))(((((....))))).---- ( -15.10, z-score = -1.80, R) >droEre2.scaffold_4929 15300277 54 - 26641161 ------GAUGCAAUUGCAAAUGCAGUCCUUGUAGUUGUAGUUGUCGUUGUCGUUGCAACG---- ------...((((((((((.(((((...))))).))))))))))((((((....))))))---- ( -16.70, z-score = -2.80, R) >droAna3.scaffold_12916 4528187 64 - 16180835 GAUGCAGUUGCAUUUUCAGAUGCAGUCGUAGUCGUUGUCGUAGUCGUAGUUGUUGCCGCUGCCA (((((..(((((((....)))))))..))).))......(((((.((((...)))).))))).. ( -16.50, z-score = -1.15, R) >droVir3.scaffold_12723 1259698 54 + 5802038 ------GAUGCAAGUGCAAGUGCAACUGCAACUGCUGCAGCUGGAGCUGCAGCUGCAGCA---- ------..((((..(((....)))..))))...((((((((((......)))))))))).---- ( -29.00, z-score = -1.94, R) >consensus ______GAUGCAAUUGCAAAUGCAGUCCUAGUAGUUGUAGUUGUAGUUGUCGUUGCAGCA____ ........((((........)))).........((((((((((......))))))))))..... (-11.35 = -11.10 + -0.25)
Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:24 2011