
| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,106,079 – 14,106,171 |
| Length | 92 |
| Max. P | 0.643045 |

| Location | 14,106,079 – 14,106,171 |
|---|---|
| Length | 92 |
| Sequences | 12 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 77.50 |
| Shannon entropy | 0.42900 |
| G+C content | 0.56740 |
| Mean single sequence MFE | -32.57 |
| Consensus MFE | -20.71 |
| Energy contribution | -20.79 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.24 |
| Structure conservation index | 0.64 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.32 |
| SVM RNA-class probability | 0.643045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
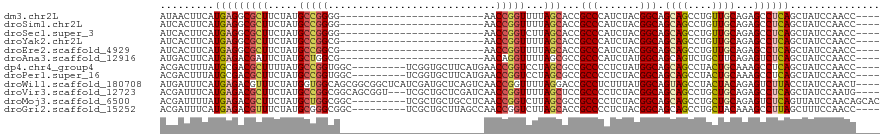
>dm3.chr2L 14106079 92 + 23011544 AUAACUUCAUGAGGCGCUUCUAUGCCGGGG------------------------AACCGGUUUUAGCACCGCCCAUCUACGGCAGCAGCCUGUUGCAGAGCCUCAGCUAUCCAACC---- .........(((((((((.....(((((..------------------------..)))))...)))........(((.((((((....)))))).)))))))))...........---- ( -32.90, z-score = -2.17, R) >droSim1.chr2L 13860545 92 + 22036055 AUCACUUCAUGAGGCGCUUCUAUGCCGGGG------------------------AACCGGUUUUAGCACCGCCCAUCUACGGCAGCAGCCUGUUGCAGAGCCUCAGCUAUCCAACC---- .........(((((((((.....(((((..------------------------..)))))...)))........(((.((((((....)))))).)))))))))...........---- ( -32.90, z-score = -2.15, R) >droSec1.super_3 364236 92 + 7220098 AUCACUUCAUGAGGCGCUUCUAUGCCGGGG------------------------AACCGGUCUUAGCACCGCCCAUCUACGGCAGCAGCCUGUUGCAGAGCCUCAGCUAUCCAACC---- .........(((((((((.....(((((..------------------------..)))))...)))........(((.((((((....)))))).)))))))))...........---- ( -32.20, z-score = -1.76, R) >droYak2.chr2L 14201950 92 + 22324452 AUCACUUCAUGAGGCGCUUCUAUGCCGGCG------------------------AACCGGUUUUAGCACCGCCCAUCUACGGCAGCAGCCUGUUGCAGAGCCUCAGCUAUCCAACC---- .........(((((((((.....(((((..------------------------..)))))...)))........(((.((((((....)))))).)))))))))...........---- ( -30.10, z-score = -1.82, R) >droEre2.scaffold_4929 15278283 92 - 26641161 AUCACUUCAUGAGGCGCUUCUAUGCCGGCG------------------------AACCGGUUUUAGCACCGCCCAUCUACGGCAGCAGCCUGUUGCAGAGCCUCAGCUAUCCAACC---- .........(((((((((.....(((((..------------------------..)))))...)))........(((.((((((....)))))).)))))))))...........---- ( -30.10, z-score = -1.82, R) >droAna3.scaffold_12916 4507422 92 - 16180835 AUGACUUCAUGAGACGAUUCUAUGCUGGCG------------------------AACAGGUUUUAGCGCCGCCCAUCUAUGGCAGCAGUCUGCUUCAGAGUCUCAGCUAUCCAACC---- .........((((((.......((((((((------------------------............))))(((.......))))))).((((...))))))))))...........---- ( -23.60, z-score = 0.05, R) >dp4.chr4_group4 1045795 107 - 6586962 ACGACUUUAUGCGACGCUUUUAUGCCGGUGGC---------UCGGUGCUUCAUGAACCGGUCCUAGCGCCGCCCCUCUAUGGCAGCAGCCUACUGCAAAGCCUCAGCUAUCCAACC---- ..........((((.(((((..(((((..((.---------.(((((((....((.....))..)))))))..))....)))))((((....))))))))).)).)).........---- ( -32.50, z-score = -0.66, R) >droPer1.super_16 1405756 107 + 1990440 ACGACUUUAUGCGACGCUUCUAUGCCGGUGGC---------UCGGUGCUUCAUGAACCGGUCCUAGCGCCGCCCCUCUACGGCAGCAGCCUACUGCAAAGCCUCAGCUAUCCAACC---- ..........((((.((((...(((((..((.---------.(((((((....((.....))..)))))))..))....)))))((((....)))).)))).)).)).........---- ( -33.30, z-score = -0.82, R) >droWil1.scaffold_180708 11171737 116 - 12563649 AUGAUUUCAUGAGACGUUUCUAUGGUGGCAGCGGCGGCUCAUCGAUGCUCAGUCAACCGGUUUUAGGACCGCCUCUUUAUGGCAGUAGCCUACUACAGAGUCUUACCUAUCCAACU---- ..(((....((((((...(((.(((((((.((((((((........)))..)))....((((....))))(((.......))).)).))).)))).)))))))))...))).....---- ( -30.80, z-score = -0.14, R) >droVir3.scaffold_12723 1228882 113 + 5802038 ACGAUUUCAUGAGACGCUUCUAUGCCGGCGGCAGCGGU---UCGCUGCUCGAUCAACCGGUUUUAGCUCCGCCCCUCUACGGCAGCAGCCUGCUGCAGAGCCUCAGCUAUCCAAUG---- ..(((....((((..(((.....(((((((((((((..---.)))))).)).....)))))...))).......((((.((((((....)))))).)))).))))...))).....---- ( -38.90, z-score = -0.79, R) >droMoj3.scaffold_6500 22200031 111 - 32352404 ACGAUUUUAUGAGACGCUUCUAUGCUGGCGGC---------UCGCUGCUGCCUCAACCGGUCUUAGCGCCGCCCCUCUACGGCAGCAGCCUGCUGCAGAGUCUCAGUUAUCCAACAGCAC ..(((....((((((((......))..(((((---------..(((((((((.....((((......)))).........)))))))))..)))))...))))))...)))......... ( -43.64, z-score = -2.73, R) >droGri2.scaffold_15252 4628122 107 + 17193109 ACGAUUUCAUGAGACGUUUCUAUGCGGGCGGC---------UCGCUGCUUAGCCAACCGGUCUUAGCACCGCCCCUCUACGGCAGCAGCCUGCUACAAAGCCUUAGCUUUCCAACC---- ..............(((......)))((((((---------(.(((((((((.....((((......)))).....))).)))))))))).)))..(((((....)))))......---- ( -29.90, z-score = -0.10, R) >consensus AUGACUUCAUGAGACGCUUCUAUGCCGGCG________________________AACCGGUUUUAGCACCGCCCAUCUACGGCAGCAGCCUGCUGCAGAGCCUCAGCUAUCCAACC____ .........(((((((((.....(((((............................)))))...)))...(((.......))).((((....))))...))))))............... (-20.71 = -20.79 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:22 2011