| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,097,222 – 14,097,333 |
| Length | 111 |
| Max. P | 0.905353 |

| Location | 14,097,222 – 14,097,333 |
|---|---|
| Length | 111 |
| Sequences | 12 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 64.63 |
| Shannon entropy | 0.76742 |
| G+C content | 0.41266 |
| Mean single sequence MFE | -28.50 |
| Consensus MFE | -9.21 |
| Energy contribution | -9.01 |
| Covariance contribution | -0.20 |
| Combinations/Pair | 1.77 |
| Mean z-score | -1.68 |
| Structure conservation index | 0.32 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.905353 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
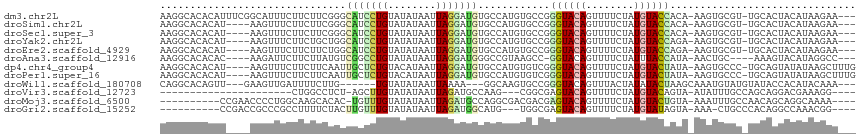
>dm3.chr2L 14097222 111 - 23011544 AAGGCACACAUUUCGGCAUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACA-AAGUGCGU-UGCACUACAUAAGAA--- ..((((((......(((.......(....)((((((((........))))))))))).)))))).((((((........))))))...-.(((((..-.))))).........--- ( -37.50, z-score = -3.58, R) >droSim1.chr2L 13851821 107 - 22036055 AAGGCACACAU----AAGUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACA-AAGUGCGU-UGCACUACAUAAGAA--- ..((((((...----(((......))).((((((((((........)))))))).)).)))))).((((((........))))))...-.(((((..-.))))).........--- ( -35.50, z-score = -3.29, R) >droSec1.super_3 355535 107 - 7220098 AAGGCACACAU----AAGUUUCUUCUUCGGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCACA-AAGUGCGU-UGCACUACAUAAGAA--- ..((((((...----(((......))).((((((((((........)))))))).)).)))))).((((((........))))))...-.(((((..-.))))).........--- ( -35.50, z-score = -3.29, R) >droYak2.chr2L 14192640 107 - 22324452 AAGGCACACAU----AAGUUUCUUCUGCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAGA-AAGUGCGU-UGCACUACAUAAGAA--- ..((((((...----.(((.......)))(((((((((........)))))..)))).)))))).((((((........))))))...-.(((((..-.))))).........--- ( -34.60, z-score = -2.46, R) >droEre2.scaffold_4929 15269421 107 + 26641161 AAGGCACACAU----AAGUUUCUUCUUCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAGA-AAGUGCGU-UGCACUACAUAAGAA--- ..((((((((.----(((......))).))((((((((........))))))))....)))))).((((((........))))))...-.(((((..-.))))).........--- ( -35.50, z-score = -3.28, R) >droAna3.scaffold_12916 4498730 103 + 16180835 AAGGCACACAC----AAGAUUCUUCUUAUGUCGGCCUGUAUAUAAUUAGGAUGGGCCGUAAGCC-GGUACAGUUUUCUAUUUACCAUA-AACUGC----AAAGUACAUAGGCC--- ..(((..(((.----((((....)))).)))((((((((.((....))..))))))))...)))-((((.(((.....)))))))...-.(((..----..))).........--- ( -24.20, z-score = -0.73, R) >dp4.chr4_group4 1035394 110 + 6586962 AAGGCACACAU----AAGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUA-AAGUGCCC-UGCAGUAUAUAAGCUUUG ..(((((....----(((......)))..........(((((((.......((((((........))))))......)))))))....-..))))).-.................. ( -22.92, z-score = 0.21, R) >droPer1.super_16 1395303 110 - 1990440 AAGGCACACAU----AAGUUUCUUCUUCAAUUGCUCUGUACAUAAUUAGGAUGUGCCAUGUGUCGGGUACAGUUUUCUAUGUACUAUA-AAGUGCCC-UGCAGUAUAUAAGCUUUG ..(((((....----(((......)))..........(((((((.......((((((........))))))......)))))))....-..))))).-.................. ( -22.92, z-score = 0.21, R) >droWil1.scaffold_180708 11158704 101 + 12563649 CAGGCACAGUU---GAAGUUGAUUUUCUUG------UGUAUAUAAUUAAAA---GGCAAGUGCCGGGUACAGUUUACUAUAUACUAAGCAAAUGUAUGUAUACCACAUACAAA--- ...((((((..---((((.....)))))))------)((((((........---(((....))).((((.....))))))))))...))...(((((((.....)))))))..--- ( -19.80, z-score = 0.03, R) >droVir3.scaffold_12723 1214818 85 - 5802038 ----------------------CUGGCCUCU-AGCUUGUAUAUAAUUAGAUGCCAAG---CGGCGAGUACAGUUUUCUAUGUACAGUA-AUAUUUGCCAGCAGGACGAAAGG---- ----------------------.((((.(((-((.(((....)))))))).)))).(---(((((((((((........)))))(...-.)..))))).))....(....).---- ( -22.00, z-score = -0.88, R) >droMoj3.scaffold_6500 22186009 100 + 32352404 ----------CCGAACCCCUGGCAAGCACAC-UGUUUGUAUAUAAUUAGAUGCCAGGCGACGACGAGUACAGUUUUCUAUGUACUGUA-AAAUUUGCCAACAGCAGGCAAAA---- ----------.....(.((((((.((....)-)(((((........))))))))))).)...((.((((((........)))))))).-...((((((.......)))))).---- ( -24.00, z-score = -1.00, R) >droGri2.scaffold_15252 4617496 97 - 17193109 ----------CCGACCGCCCGCCUUUUCUACUUGUUUGUAUAUAAUUAGAUGGCAUG---UGGCGAGUACAGUUUUCUAUGUAUAGUA-AAA-CUGCCCACAGGCCAAACGG---- ----------(((...(((.(((...((((.((((......)))).)))).))).((---(((.(....((((((((((....))).)-)))-))))))))))))....)))---- ( -27.60, z-score = -2.17, R) >consensus AAGGCACACAU____AAGUUUCUUCUUCUGGCAUCCUGUAUAUAAUUAGGAUGUGCCAUGUGCCGGGUACAGUUUUCUAUGUACCAUA_AAGUGCGU_UGCACUACAUAAGAA___ ...............................(((((((........)))))))............((((((........))))))............................... ( -9.21 = -9.01 + -0.20)

Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:21 2011