| Sequence ID | dm3.chr2L |
|---|---|
| Location | 14,071,317 – 14,071,463 |
| Length | 146 |
| Max. P | 0.731668 |

| Location | 14,071,317 – 14,071,463 |
|---|---|
| Length | 146 |
| Sequences | 7 |
| Columns | 146 |
| Reading direction | forward |
| Mean pairwise identity | 79.10 |
| Shannon entropy | 0.42394 |
| G+C content | 0.47136 |
| Mean single sequence MFE | -46.00 |
| Consensus MFE | -24.47 |
| Energy contribution | -27.19 |
| Covariance contribution | 2.72 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.78 |
| Structure conservation index | 0.53 |
| Background model | dinucleotide |
| Decision model | sequence based alignment quality |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.731668 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
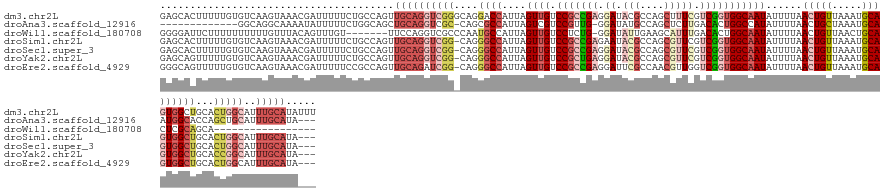
>dm3.chr2L 14071317 146 + 23011544 GAGCACUUUUUGUGUCAAGUAAACGAUUUUUCUGCCAGUUGCAGGUCGGGCAGGACCAUUAGUUGUCCGCCGAGGAUACGCCAGCUUUCGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACUGGCAUUUGCAUAUUU ((.(((.....)))))..(((((.((....))(((((((.(((((((......))))....((((.(((((((.((...........)).)))))))))))......(((((.....)))))...)))))))))))))))...... ( -48.40, z-score = -1.93, R) >droAna3.scaffold_12916 4471168 128 - 16180835 -------------GGCAGGCAAAAUAUUUUUCUGGCAGCUGCAGGUCGC-CAGCGCCAUUAGUCGUCCGUUG-GGAUAUGCCAGCUCUUGACACUGGCCAUAUUUUAACUGCUAAAUGCAAUGGCACCAGCUGCAUUUGCAUA--- -------------(((((..(((((((....(((((.((.....)).))-))).((((...((((...((((-(......)))))...))))..)))).)))))))..)))))..((((((..(((.....)))..)))))).--- ( -44.10, z-score = -0.99, R) >droWil1.scaffold_180708 11119440 121 - 12563649 GGGGAUUCUUUUUUUUUUGUUUACAGUUUGU-------UUCCAGGUCGCCCAAUGCCAUUAGUUGUCCUCUG-GGAUAUUGAAGCAUUUGACACUGGCAAUAUUUUAACUGUUAACUGCACUCGCAGCA----------------- ..................(((.(((((((((-------(.((((((((...(((((..(((((.((((....-))))))))).))))))))).)))).))))....)))))).)))(((....)))...----------------- ( -29.60, z-score = -1.01, R) >droSim1.chr2L 13825832 142 + 22036055 GAGCACUUUUUGUGUCAAGUAAACGAUUUUUCUGCCAGUUGCAGGUCGG-CAGGGCCAUUAGUUGUCCGCCGAGAAUACGCCAGCGUUCGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACUGGCAUUUGCAUA--- ((.(((.....)))))..(((((.((....))(((((((.((((.((((-(.((((........)))))))))......((((.((.....)).)))).........(((((.....)))))..))))))))))))))))...--- ( -51.10, z-score = -2.60, R) >droSec1.super_3 329387 142 + 7220098 GAGCACUUUUUGUGUCAAGUAAACGAUUUUUCUGCCAGUUGCAGGUCGG-CAGGGCCAUUAGUUGUCCGCCGAGGAUACGCCAGCGUUCGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACUGGCAUUUGCAUA--- ((.(((.....)))))..(((((.((....))(((((((.((((.((((-(...)))....((((.(((((((.((.(((....))))).)))))))))))......(((((.....)))))))))))))))))))))))...--- ( -53.00, z-score = -2.92, R) >droYak2.chr2L 14165209 142 + 22324452 GAGCAGUUUUUGUGUCAAGUAAACGAUUUUUCUGCCAGUUGCAGGUCGG-CAGGGCCAUUAGUUGUCCGCUGAGGAUACGCCAGCGUUCGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACCGGCAUUUGCAUA--- (.((((...((((.........)))).....)))))...((((.(((((-...((((....((((.(((((((.((.(((....))))).)))))))))))......(((((.....)))))))))...)))))...))))..--- ( -48.70, z-score = -1.75, R) >droEre2.scaffold_4929 15243292 142 - 26641161 GGGCAGUUUUUGUGUCAAGUAAACGAUUUUUCCGCCAGUUGCAGAUCGG-CAGGGCCAUUAGUUGUCCGCCGAGGAUUCGCCAACGUUGGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACUGGCAUUUGCAUA--- .((((.......))))..(((((.((....)).((((((.((((.((((-(.((((........)))))))))......((((.((.....)).)))).........(((((.....)))))..)))))))))).)))))...--- ( -47.10, z-score = -1.27, R) >consensus GAGCACUUUUUGUGUCAAGUAAACGAUUUUUCUGCCAGUUGCAGGUCGG_CAGGGCCAUUAGUUGUCCGCCGAGGAUACGCCAGCGUUCGUCGGUGGCAAUAUUUUAACUGUUAAAUGCAGUGGCUGCACUGGCAUUUGCAUA___ .......................................((((.(((((....((((....((((.(((((((.((.(((....))))).)))))))))))......(((((.....)))))))))...)))))...))))..... (-24.47 = -27.19 + 2.72)


Generated by rnazCluster.pl (part of RNAz 1.0) on Tue Apr 19 21:39:18 2011